Genetic structure of populations and introgression of three Quercus species in mountainous area of Beijing
-
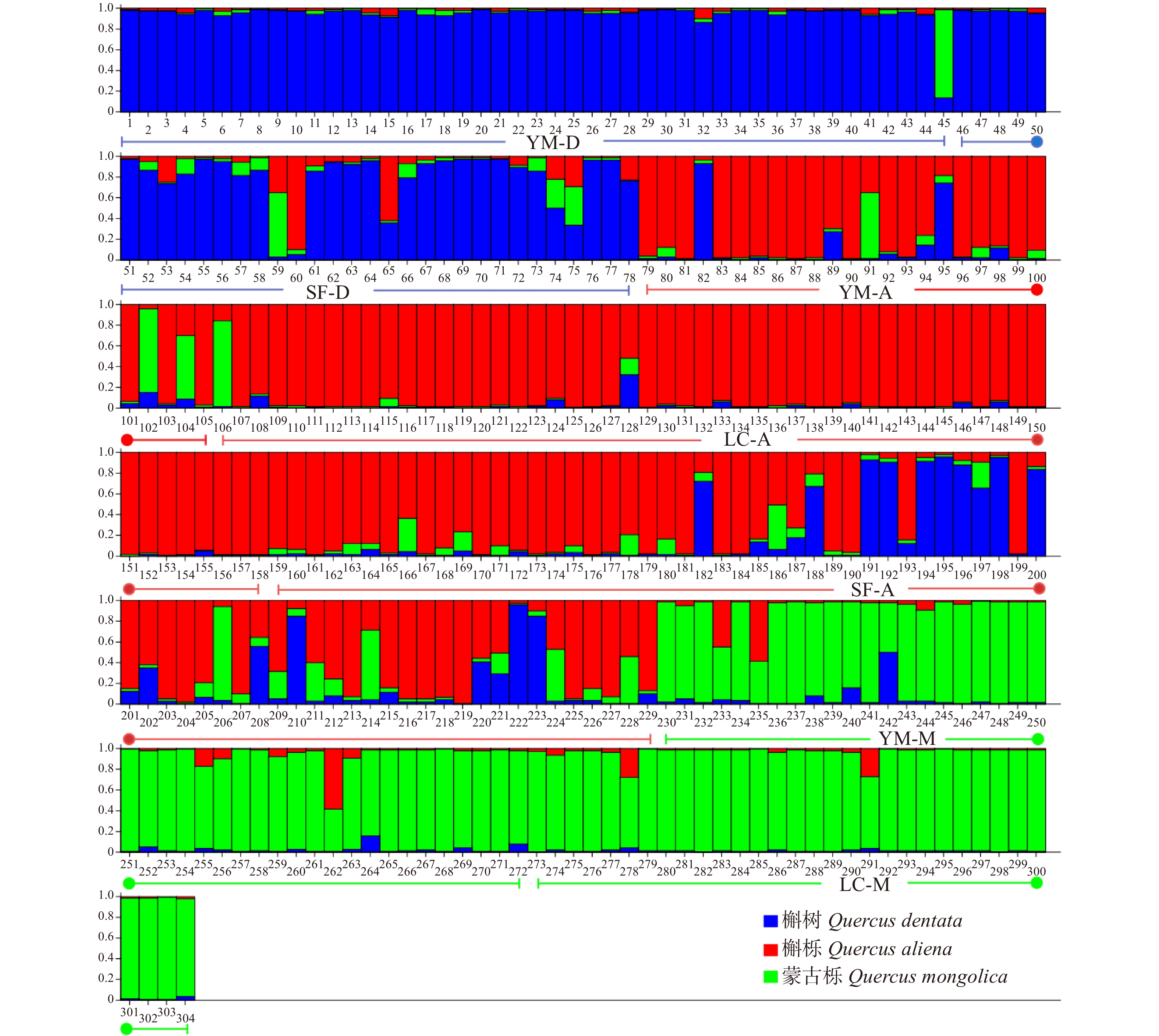
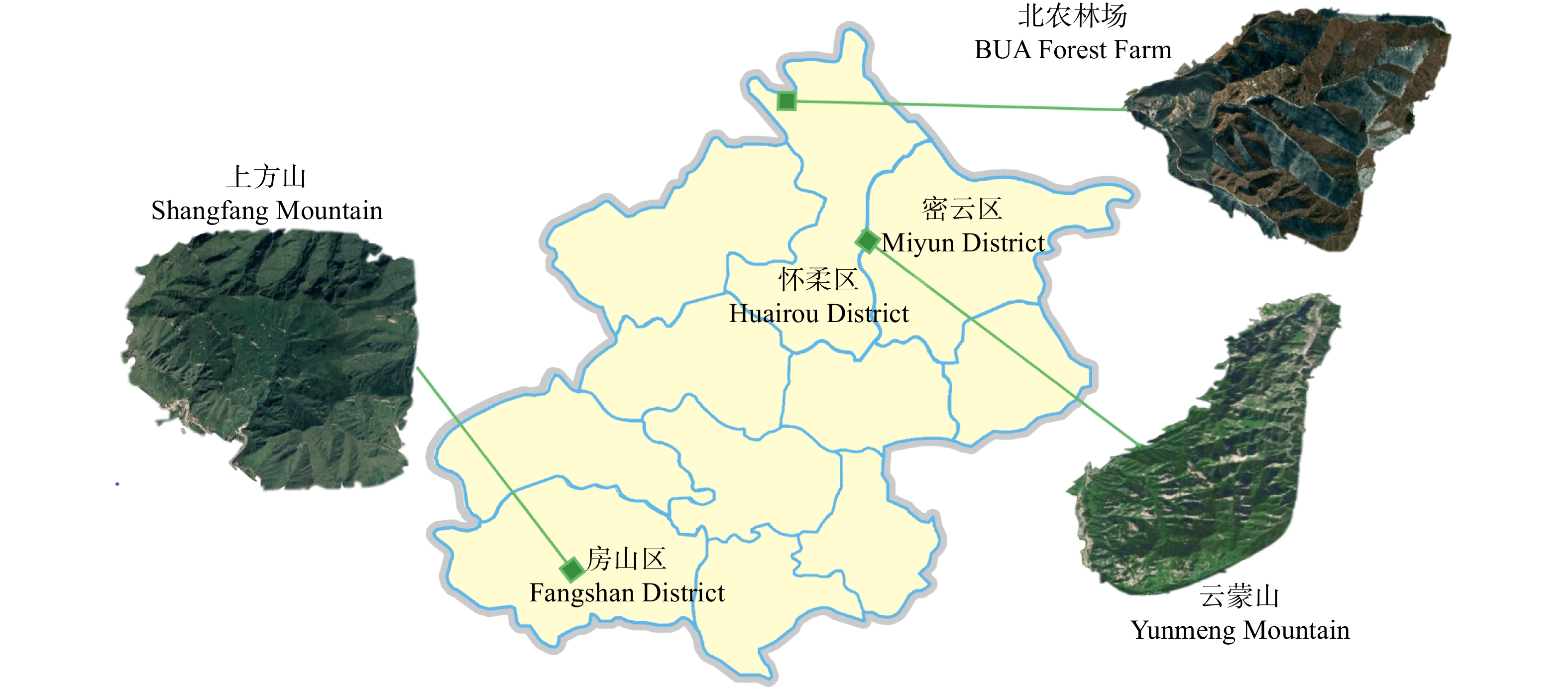
摘要:目的 栎属植物种间经常发生种间杂交和基因渐渗现象,特别是同域分布的同组内栎树之间,这种情况会更加频繁。本文通过对北京山区3种栎属植物居群遗传结构遗传变异与遗传结构进行研究,为了解北京地区自然分布的栎属植物种间基因渐渗情况、种质资源现状以及经营管理提供有效数据。方法 本文使用6对SSR引物对云蒙山、上方山和北农林场同域分布的304个蒙古栎、槲树、槲栎的居群遗传多样性、遗传结构和种间基因渐渗进行了研究。结果 共检测到等位标记105个,每个位点的平均等位标记数(Na)为17.5个,期望杂合度(He)为0.660 ~ 0.911,平均为0.838,多态性信息含量指数(PIC)为0.632 ~ 0.903,平均为0.822,3种栎树在总体水平上具有较高的遗传多样性。在种的水平上,3种栎树的平均等位标记数(Na)为12.667 ~ 14.167,期望杂合度(He)为0.743 ~ 0.849,多态性信息含量指数(PIC)为0.725 ~ 0.826,3种栎属植物的遗传多样性水平为蒙古栎 > 槲树 > 槲栎。对7个栎树居群的遗传结构分析表明,遗传变异大部分发生在居群内。通过Structure软件对3种栎树种间基因渐渗进行分析,发现槲树−槲栎、槲树−蒙古栎和槲栎−蒙古栎这3个种对间均有基因渐渗发生。结论 在北京山区分布的蒙古栎、槲树和槲栎这3种栎属植物之间存在普遍的、复杂的渐渗杂交现象。Abstract:Objective Interspecific hybridization and introgression often occur among species of Quercus, especially among sympatric Quercus more frequently. In this paper, the genetic structure and genetic variation of three species of Quercus in the mountainous area of Beijing were studied. It will provide effective data for gene introgression, germplasm resources and management of Quercus species naturally distributed in Beijing.Method 304 natural oak samples of Quercus mongolica, Quercus dentate and Quercus aliena from Yunmeng Mountain, Shangfang Mountain and BUA (Beijing University of Agriculture) Forest Farm were selected as the material, and six pairs of SSR primers were used to study the genetic diversity, genetic structure and introgressive hybridization among populations.Result A total of 105 alleles were detected, the average number of alleles (Na) of each locus was 17.5, the expected heterozygosity (He) was 0.660−0.911, the polymorphism information content index (PIC) was 0.632−0.903. The three oaks had high genetic diversity at the overall level. At the species level, the average allele number (Na) of the three oaks was 12.667−14.167, the expected heterozygosity (He) was 0.743−0.849, the polymorphism information content index (PIC) was 0.725−0.826, the genetic diversity level of the three species of oaks was Q. mongolica > Q. dentate > Q. aliena. The genetic structure analysis of 7 populations of Quercus showed that most of the genetic variation was in local populations. The genetic introgression analysis by structure software found that the gene introgression occurred among three pairs of Q. mongolica, Q. dentate and Q. aliena.Conclusion There is a common and complex phenomenon of introgression hybridization among Q. mongolica, Q. dentate and Q. aliena in Beijing mountainous area.
-
Keywords:
- Quercus /
- genetic diversity /
- genetic structure /
- gene introgression
-
-
表 1 6对SSR引物多态性
Table 1 Ploymorphism of 6 SSR primers
位点
Locus引物(5′—3′)
Primer(5′−3′)重复片段
Repeat motif等位标记数
Number of alleles(Na)观测杂合度
Observation heterozygosity
(Ho)期望杂合度
Expected heterozygosity
(He)多态信息量
Polymorphism information content(PIC)Qden 03011 AACCCAACCTTCCCTTCATC (AG)8 15.0 0.823 0.836 0.817 GCAGTGGTGCCTAATGTAGAC Qden 05011 CCCACTCCCTGTCCATTGT (CT)8 19.0 0.881 0.893 0.882 CACTGTGTGCTGCGACTTG Qden 03032 AGTTGTGGTCCTGCTCGC (CT)12 16.0 0.618 0.857 0.841 GAAAAGTGCGATGACGGTTG Qden 05031 CCCCGATTCGCCATCATTGT (GT)12 19.0 0.846 0.873 0.859 GTAACGCCGTTTTTCTCCACC ssrQpZAG112 TTCTTGCTTTGGTGCGCG (GA)32 11.0 0.485 0.660 0.632 GTGGTCAGAGACTCGGTAAGTATTC ssrQpZAG96 CCCAGTCACATCCACTACTGTCC (TC)20 25.0 0.881 0.911 0.903 GGTTGGGAAAAGGAGATCAGA 平均值 Mean 17.5 0.755 0.838 0.822 表 2 7个栎树居群的遗传多样性
Table 2 Genetic diversity of 7 populations of oak
物种
Speices居群
Population样本数
Sample numberNa Ho He 近交系数
Inbreeding coefficient(F)PIC 槲树
Quercus dentateYM-D 45 9.833 0.819 0.762 − 0.086 0.718 SF-D 33 10.833 0.758 0.797 0.036 0.760 平均值 Mean 78 12.667 0.793 0.787 − 0.014 0.754 槲栎
Quercus alienaYM-A 27 7.833 0.685 0.671 − 0.027 0.629 LC-A 53 8.000 0.657 0.683 0.063 0.650 SF-A 71 12.833 0.753 0.771 0.007 0.750 平均值 Mean 151 14.167 0.706 0.743 0.046 0.725 蒙古栎
Quercus mongolicaYM-M 43 11.667 0.797 0.831 0.022 0.800 LC-M 32 9.833 0.854 0.839 − 0.031 0.806 平均值 Mean 75 13.167 0.826 0.849 0.016 0.826 注:YM. 云蒙山样地,SF. 上方山样地,LC.北农林场样地,D.槲树,A.槲栎,M.蒙古栎。Notes: YM refers to Yunmeng Mountain sample plot; SF refers to Shangfang Mountain sample plot; LC refers to BUA Forest Farm sample plot;D refers to Quercus dentate; A refers to Quercus aliena; M refers to Quercus mongolica. 表 3 同种栎树居群间遗传变异和基因流
Table 3 Genetic variation and Nm values between populations within same oak species
居群1
Population 1居群2
Population 2基因分化系数
Genetic differentiation coefficient(Fst)基因流
Gene flow(Nm)YM-D SF-D 0.019 12.710 YM-A LC-A 0.044 5.472 YM-A SF-A 0.032 7.649 LC-A SF-A 0.020 12.027 YM-M LC-M 0.025 9.924 表 4 异种栎树居群间遗传变异和基因流
Table 4 Genetic variation and Nm values between populations among varied oak species
居群1 Population 1 居群2 Population 2 Fst Nm YM-D YM-A 0.101 2.237 YM-D YM-M 0.070 3.342 YM-A YM-M 0.093 2.445 LC-A LC-M 0.108 2.068 SF-D SF-A 0.045 5.271 表 5 居群间遗传距离
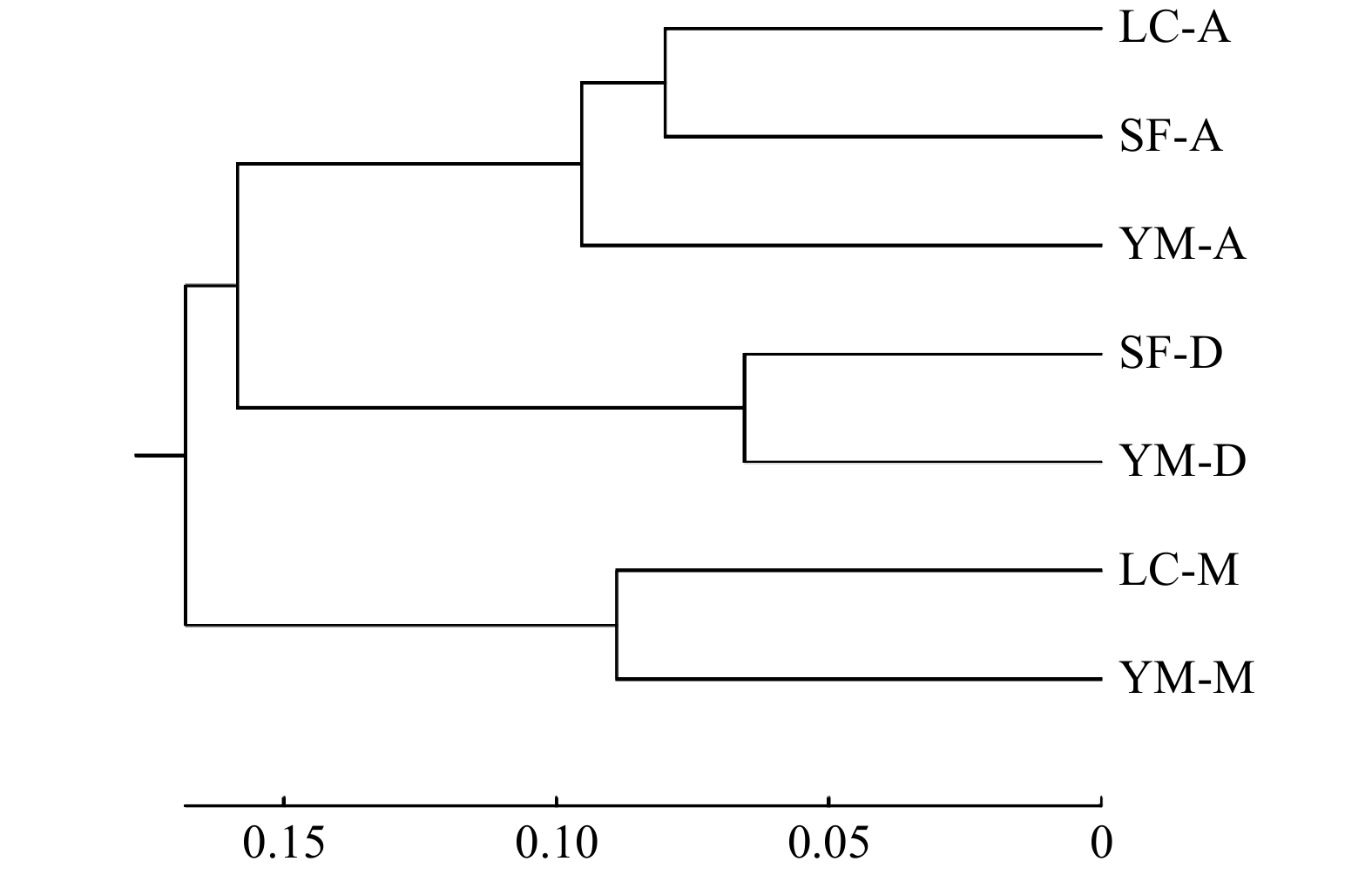
Table 5 Genetic distance between populations
居群 Population LC-A LC-M SF-A SF-D YM-A YM-D YM-M LC-A — LC-M 0.427 — SF-A 0.160 0.285 — SF-D 0.374 0.343 0.196 — YM-A 0.218 0.368 0.164 0.286 — YM-D 0.448 0.394 0.264 0.131 0.334 — YM-M 0.400 0.178 0.231 0.268 0.305 0.340 — -
[1] Anderson E, Hubricht L. Hybridization in Tradescantia (Ⅲ): the ecidence for introgressive for introgressive hybridization[J]. American Journal of Botany, 1938, 25(6): 396−402. doi: 10.1002/j.1537-2197.1938.tb09237.x
[2] Rieseberg L H, Wendel J F. Introgression and its consequences in plants [M]// Harrison R. Hybrid zones and the evolutionary process. Oxford: Oxford University Press, 1993: 70−103.
[3] Rushton B S. Natural hybridization within the genus Quercus L.[J]. Annales des Sciences Forestieres, 1993, 50 (Suppl.): 73−90.
[4] Arnold M L. Evolution through genetic exchange[M]. Oxford: Oxford University Press, 2006.
[5] Jiggins C D, Mallet J. Bimodal hybrid zones and speciation[J]. Trends in Ecology & Evolution, 2000, 15(6): 250−255.
[6] Hamrick J L. Plant population genetics, breeding and genetic resources[M]. Sunderland: Sinauer Associates, 1990.
[7] Kremer A, Petit R. Gene diversity in natural populations of oak species[J]. Annales des Sciences Forestières, 1993, 50: 186−202. doi: 10.1051/forest:19930717
[8] Quang N D, Ikeda S, Harada K. Nucleotide variation in Quercus crispula Blume[J]. Heredity, 2008, 101(2): 166−174. doi: 10.1038/hdy.2008.42
[9] Arnold M L, Ballerini E S, Brothers A N. Hybrid fitness, adaptation and evolutionary diversification: lessons learned from Louisiana irises[J]. Heredity, 2012, 108(3): 159−166. doi: 10.1038/hdy.2011.65
[10] Eaton D A, Hipp A L, González-Rodríguez A, et al. Historical introgression among the American live oaks and the comparative nature of tests for introgression[J]. Evolution, 2015, 69(10): 2587−2601.
[11] Dow B D, Ashley M V. High levels of gene flow in bur oak revealed by paternity analysis using microsatellites[J]. Journal of Heredity, 1998, 89(1): 62−70. doi: 10.1093/jhered/89.1.62
[12] Muir G, Fleming C C, Schlotterer C, et al. Species status of hybridizing oaks[J]. Nature, 2000, 405: 1016. doi: 10.1038/35016640
[13] Steinhoff S. Results of species hybridization with Quercus robur L. and Quercus petraea (Matt) Liebl[J]. Annales des Sciences Forestieres, 1993, 50 (Suppl.): 137−143.
[14] Lefort E, Lally M, Thompson D. Morphological traits, microsatellite fingerprinting and genetic relatedness of a stand of elite oaks (Q. robur L.) at Tullynally Ireland[J]. Silvae Genetica, 1998, 473(176): 5−6.
[15] Lopez-Aljorna A, Angeles B M, Aguinagalde I, et al. Fingerprinting and genetic variability in cork oak (Quercus suber L.) elite trees using ISSR and SSR markers[J]. Annales of Sciences Forestieres, 2007, 64(7): 773−779.
[16] Moran E V, Willis J, Clark J S. Genetic evidence for hybridization in red oaks (Quercus Sect. Lobatae, Fagaceae)[J]. American Journal of Botany, 2012, 99(1): 92−100. doi: 10.3732/ajb.1100023
[17] Petit R J, Csaikl U M, Bordács S, et al. Chloroplast DNA variation in European white oaks[J]. Forest Ecology and Management, 2002, 156(1): 5−26.
[18] Antonecchia G, Fortini P, Lepais O, et al. Genetic structure of a natural oak community in central Italy: evidence of gene flow between three sympatric white oak species (Quercus, Fagaceae)[J]. Annals of Forest Research, 2015, 57(2): 205−216.
[19] Salvini D, Bruschi P, Fineschi S, et al. Natural hybridisation between Quercus petraea (Matt.) Liebl. and Quercus pubescens Willd. within an Italian stand as revealed by microsatellite fingerprinting[J]. Plant Biology, 2009, 11(5): 758−765. doi: 10.1111/j.1438-8677.2008.00158.x
[20] Lepais O, Pettt R J, Guichoux E, et al. Species relative abundance and direction of introgression in oaks[J]. Molecular Ecology, 2009, 18(10): 2228−2242. doi: 10.1111/j.1365-294X.2009.04137.x
[21] 厉月桥, 李迎超, 吴志庄. 中国北方栎属植物资源调查与区划[J]. 林业资源管理, 2013(4):88−93. doi: 10.3969/j.issn.1002-6622.2013.04.017 Li Y Q, Li Y C, Wu Z Z. Study on investigation and division of the resources of Quercus in northern China[J]. Forest Resources Management, 2013(4): 88−93. doi: 10.3969/j.issn.1002-6622.2013.04.017
[22] 李文英, 顾万春, 周世良. 蒙古栎天然群体遗传多样性的AFLP分析[J]. 林业科学, 2003, 39(5):29−36. doi: 10.3321/j.issn:1001-7488.2003.05.005 Li W Y, Gu W C, Zhou S L. AFLP analysis on genetic diversity of Quercus mongolica populations[J]. Scientia Silvae Sinicae, 2003, 39(5): 29−36. doi: 10.3321/j.issn:1001-7488.2003.05.005
[23] 徐小林, 徐立安, 黄敏仁, 等. 栓皮栎天然群体SSR遗传多样性研究[J]. 遗传, 2004, 26(5):683−688. doi: 10.3321/j.issn:0253-9772.2004.05.023 Xu X L, Xu L A, Huang M R, et al. Genetic diversity of microsatellites (SSRs) of natural populations of Quercus variabilis[J]. Hereditas (Beijing), 2004, 26(5): 683−688. doi: 10.3321/j.issn:0253-9772.2004.05.023
[24] 魏高明. 苏皖4种同域分布栎树的遗传变异与基因渐渗[D]. 南京: 南京林业大学, 2015. Wei G M. Genetic variation of populations and introgression among four sympatric oaks in Jiangsu and Anhui provinces[D]. Nanjing: Nanjing Forestry University, 2015.
[25] Zeng Y F, Liao W J, Petit R J, et al. Geographic variation in the structure of oak hybrid zones provides insights into the dynamics of speciation[J]. Molecular Ecology, 2011, 20(23): 4995−5011. doi: 10.1111/j.1365-294X.2011.05354.x
[26] Hubert F, Grimm G W, Jousselin E, et al. Multiple nuclear genes stableilize the phylogenetic backbone of the genus Quercus[J]. Systematics & Biodiversity, 2014, 12(4): 405−423.
[27] 任宪威. 北京新植物[J]. 河北农业大学学报, 1996, 19(3):86−87. Ren X W. New taxa from Beijing[J]. Journal of Agricultural University of Hebei, 1996, 19(3): 86−87.
[28] 陈焕镛, 黄成就.中国植物志(22): 壳斗科[M]. 北京: 科学出版社, 1998: 213−263. Chen H Y, Huang C J. Flora of China (22): Fagaceae [M]. Beijing: Science Press, 1998: 213−263.
[29] Kampfer S, Lexer C, Steinkellner H, et al. Characterization of (GA)n microsatellite loci from Quercus robur[J]. Hereditas, 1998, 129: 183−186.
[30] Aldrich P R, Michler C H, Sun W L, et al. Microsatellite markers for northern red oak (Fagaceae: Quercus rubra)[J]. Molecular Ecology Notes, 2002, 2: 472−474. doi: 10.1046/j.1471-8286.2002.00282.x
[31] Steinkellner H, Fluch S, Turetschek E, et al. Identification and characterization of (GA / CT)n-microsatellite loci from Quercus petraea[J]. Plant Molecular Biology, 1997, 33: 1093−1096. doi: 10.1023/A:1005736722794
[32] 王越. 基于SSR标记的槲树、蒙古-辽东栎种间杂交研究[D]. 济南: 山东大学, 2012. Wang Y. Natural hybridization between Quercus dentata and Q. mongolica-liaotungensis revealed by microsatellite markers[D]. Jinan: Shandong University, 2012.
[33] Marshall T C, Slate J, Kruuk L E B, et al. Statistical confidence for likelihood-based paternity inference in natural populations[J]. Molecular Ecology, 1998, 7(5): 639−655. doi: 10.1046/j.1365-294x.1998.00374.x
[34] Peakall R, Smouse P E. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research: an update[J]. Bioinformatics, 2012, 28(19): 2537−2539. doi: 10.1093/bioinformatics/bts460
[35] 范英明, 张登荣, 于大德, 等. 河北省华北落叶松天然群体遗传多样性分析[J]. 植物遗传资源学报, 2014, 15(3):465−471. Fan Y M, Zhang D R, Yu D D, et al. Genetic diversity and population structure of Larix principis-rupprechtii Mayr in Hebei Province[J]. Journal of Plant Genetic Resources, 2014, 15(3): 465−471.
[36] 张如华. 柽柳群体遗传变异研究[D]. 南京: 南京林业大学, 2011. Zhang R H. Study on the gentic variation of Tamarix chinensis Lour. populations [D]. Nanjing: Nanjing Forestry University, 2011.
[37] 张学江. 中国卧龙自然保护区不同海拔川滇高山栎(Quercus aquifolioides)群体的遗传变异[D]. 成都: 中国科学院成都生物研究所2006. Zhang X J. Genetic variation of Quercus aquifolioides populations at varying altitudes in the Wolong Nature Reserve of China[D]. Chengdu: Chengdu Institute of Biology, 2006.
[38] Hardy O J. Fine-scale genetic structure and gene dispersal in Centaurea corymbosa (Asteraceae) (Ⅱ): correlated paternity within and among sibships[J]. Genetics, 2004, 168(3): 1601−1614. doi: 10.1534/genetics.104.027714
[39] Craft K J, Ashley M V. Landscape genetic structure of bur oak (Quercus macrocarpa) savannas in Illinois[J]. Forest Ecology and Management, 2007, 239(1): 13−20.
[40] 邸晓瑶. 基于cpDNA和SSR标记的槲栎群体遗传学研究[D]. 西安: 西北大学, 2017. Di X Y. Population genetics of Quercus aliena based on cpDNA and SSR marker[D]. Xi’an: Northwest University, 2017.
[41] Chybicki I J, Burczyk J. Seeing the forest through the trees: comprehensive inference on individual mating patterns in a mixed stand of Quercus robur and Q. petraea[J]. Annals of Botany, 2013, 112(3): 561−574. doi: 10.1093/aob/mct131
[42] 徐刚标.植物群体遗传学[M]. 北京: 科学出版社, 2009: 55−65. Xu G B. Plant population genetics[M]. Beijing: Science Press, 2009: 55−65.
[43] Liu Y, Li Y, Song J, et al. Geometric morphometric analyses of leaf shapes in two sympatric Chinese oaks: Quercus dentata Thunberg and Quercus aliena Blume (Fagaceae)[J/OL]. Annals of Forest Science, 2018, 75(4)[2019−08−21]. http://link.springer.com/article/10.1007/s13595-018-0770-2.
[44] 解新明, 云锦凤. 植物遗传多样性及其检测方法[J]. 中国草地, 2000, 22(6):52−60. Xie X M, Yun J F. Genetic diversity and detective methods of plant[J]. Chinese Journal of Grassland, 2000, 22(6): 52−60.
[45] 鲜冬娅. 北京上方山植物多样性及保护研究[D]. 北京: 北京林业大学, 2008. Xian D Y. Study on plant diversity and conservation in Shangfang Mountain, Beijing[D]. Beijing: Beijing Forestry University, 2008.
[46] Aldrich P R, Lavender-Bares J. Wild crop relatives: genomic and breeding resources[M]. Berlin: Springer Berlin Heidelberg, 2011.
[47] Burgarella C, Lorenzo Z, Jabbour-Zahab R, et al. Detection of hybrids in nature: application to oaks (Quercus suber and Q. ilex)[J]. Heredity, 2009, 102(5): 442−452. doi: 10.1038/hdy.2009.8
[48] Curtu A L, Gailing O, Finkeldey R. Evidence for hybridization and introgression within a species-rich oak (Quercus spp.) community [J/OL]. BMC Evolutionary Biology, 2007, 7(1): 218 [2019−08−21].http://bmcevolbiol.biomedcentral.com/articles/10.1186/1471-2148-7-218.
[49] Lyu J, Song J, Liu Y, et al. Species boundaries between three sympatric oak species: Quercus aliena, Q. dentata, and Q. variabilis at the northern edge of their distribution in China[J/OL]. Frontiers in Plant Science, 2018, 9: 414[2019−06−14]. http://www.frontiersin.org/articles/10.3389/fpls.2018.00414/full.
-
期刊类型引用(24)
1. 张秀芸,伍文慧,梁英梅. 落叶松枯梢病在中国的适生性. 生态学报. 2024(07): 3027-3037 .  百度学术
百度学术
2. 葛婉婷,刘莹,赵智佳,张珅,李洁,杨桂娟,曲冠证,王军辉,麻文俊. 不同气候情景下黄心梓木在我国的潜在适生区预测. 林业科学. 2024(11): 63-74 .  百度学术
百度学术
3. 汤思琦,武扬,梁定东,郭恺. 未来气候变化下栎树猝死病菌在中国的适生性分析. 生态学报. 2023(01): 388-397 .  百度学术
百度学术
4. 刘璐璐,赵亮,蔺诗颖,冯建龙. 基于MaxEnt和GARP的阿蒙森海域南极磷虾(EUPHAUSIA SUPERBA)的分布区预测. 海洋与湖沼. 2023(02): 399-411 .  百度学术
百度学术
5. 唐雨薇,张晓龙,张雪云,吕佩锋,罗乐. 基于GIS与AHP分析法的单叶蔷薇生态适宜性评价. 绿色科技. 2023(13): 205-208+213 .  百度学术
百度学术
6. 王广平,李成,王书砚,刘超,杨君珑. 宁夏罗山青海云杉林空间分布特征研究. 农业科学研究. 2023(03): 10-15+23 .  百度学术
百度学术
7. 张惠惠,孟祥霄,林余霖,陈士林,黄林芳. 基于GMPGIS系统和MaxEnt模型预测人参全球潜在生长区域. 中国中药杂志. 2023(18): 4959-4966 .  百度学术
百度学术
8. 李盼畔,何旭诺,吴海荣,陈萍,刘明航,武目涛,王亚锋. 多年生豚草在中国的潜在分布预测. 植物检疫. 2022(04): 57-62 .  百度学术
百度学术
9. 李盼畔,何旭诺,左然玲,吕文刚,吴海荣. 4种蒺藜草属杂草在中国的潜在适生性预测. 杂草学报. 2022(02): 15-23 .  百度学术
百度学术
10. 林姗,陆兴利,王茹琳,李庆,王明田,郭翔,文刚. RCP8.5情景下气候变化对四川省猕猴桃溃疡病病菌地理分布的影响. 江苏农业科学. 2020(03): 124-129 .  百度学术
百度学术
11. 王华辰,朱弘,李涌福,伊贤贵,李蒙,南程慧,王贤荣. 中国特有植物雪落樱桃潜在分布及其生态特征. 热带亚热带植物学报. 2020(02): 136-144 .  百度学术
百度学术
12. 赵金鹏,王茹琳,刘原,陆兴利,王庆,郭翔,文刚,李庆. RCP4.5情景下四川省猕猴桃溃疡病菌适生性分析. 沙漠与绿洲气象. 2020(02): 137-143 .  百度学术
百度学术
13. 段义忠,王佳豪,王驰,王海涛,杜忠毓. 未来气候变化下西北干旱区4种扁桃亚属植物潜在适生区分析. 生态学杂志. 2020(07): 2193-2204 .  百度学术
百度学术
14. 陈爱莉,赵志华,龚伟,孔芬,张克亮. 气候变化背景下紫楠在中国的适宜分布区模拟. 热带亚热带植物学报. 2020(05): 435-444 .  百度学术
百度学术
15. 文雪梅,艾科拜尔·木哈塔尔,木巴来克·阿布都许科尔,阿不都拉·阿巴斯. 基于MaxEnt模型的新疆微孢衣属地衣生境适宜性评价. 武汉大学学报(理学版). 2019(01): 77-84 .  百度学术
百度学术
16. 常红,刘彤,王大伟,纪孝儒. 气候变化下中国西北干旱区梭梭(Haloxylon ammodendron)潜在分布. 中国沙漠. 2019(01): 110-118 .  百度学术
百度学术
17. 吕汝丹,何健,刘慧杰,姚敏,程瑾,谢磊. 羽叶铁线莲的分布区与生态位模型分析. 北京林业大学学报. 2019(02): 70-79 .  本站查看
本站查看
18. 陆兴利,罗伟,李庆,林姗,王茹琳,游超,郭翔,王明田. RCP2.6情景下四川省猕猴桃溃疡病菌潜在分布预测. 湖北农业科学. 2019(18): 49-54 .  百度学术
百度学术
19. 王蕾,罗磊,刘平,侯晓臣,邱琴,高亚琪,李曦光. 基于MaxEnt模型分析新疆特色林果区春尺蠖发生风险. 新疆农业科学. 2019(09): 1691-1700 .  百度学术
百度学术
20. 王茹琳,郭翔,李庆,王明田,游超. 四川省猕猴桃溃疡病潜在分布预测及适生区域划分. 应用生态学报. 2019(12): 4222-4230 .  百度学术
百度学术
21. 赵健,李志鹏,张华纬,陈宏,翁启勇. 基于MaxEnt模型和GIS技术的烟粉虱适生区预测. 植物保护学报. 2019(06): 1292-1300 .  百度学术
百度学术
22. 王奕晨,郑鹏,潘文斌. 运用GARP生态位模型预测福寿螺在中国的潜在适生区. 福建农林大学学报(自然科学版). 2018(01): 21-25 .  百度学术
百度学术
23. 邱靖,朱弘,陈昕,汤庚国. 基于DIVA-GIS的水榆花楸适生区模拟及生态特征. 北京林业大学学报. 2018(09): 25-32 .  本站查看
本站查看
24. 王野,陈磊,白云,张俊娥,刘红霞,田呈明. 云杉矮槲寄生遗传多样性的ISSR分析. 西北植物学报. 2017(11): 2153-2162 .  百度学术
百度学术
其他类型引用(20)



 下载:
下载: