Identification of aspartic acid protease PtoAED3-interacting proteins through GST pull-down assays in Populus tomentosa
-
摘要:目的 天冬氨酸蛋白酶属于蛋白水解酶家族,为了解析毛白杨天冬氨酸蛋白酶PtoAED3在植物生长发育中的分子调节机制,利用GST-pull down联合质谱技术,对PtoAED3的互作蛋白进行鉴定和分析。方法 通过同源克隆获得了毛白杨PtoAED3的CDS序列,构建含GST标签的原核表达载体pGEX-4T-PtoAED3。使用IPTG诱导GST-PtoAED3融合蛋白表达后,利用GST标签对PtoAED3蛋白进行纯化,纯化后的PtoAED3蛋白与毛白杨植株总蛋白共孵育,应用GST-pull down技术获得毛白杨总蛋白中与PtoAED3蛋白相互作用的候选蛋白,随后通过质谱技术对筛选到的候选蛋白进行鉴定与分析。结果 通过质谱技术鉴定这些蛋白的氨基酸序列,共筛选出128 个与PtoAED3特异性结合的候选互作蛋白,这些互作蛋白涉及到细胞进程、代谢过程、应激反应、生物调节、发育过程等多个生物学过程。结论 通过GST-pull down实验联合质谱技术,筛选出毛白杨中与PtoAED3互作的候选蛋白,为研究毛白杨PtoAED3与底物或复合物的互作及其影响毛白杨生长发育的分子调节机制提供了初步方向。
-
关键词:
- PtoAED3 /
- GST-pull down /
- 质谱 /
- 互作蛋白
Abstract:Objective Aspartic acid protease belongs to proteolytic enzyme family. In order to further analyze the molecular regulation mechanism of PtoAED3 in plant growth and development, GST-pull down combined mass spectrometry technology was used to identify and analyze the interacting protein of protein PtoAED3 in Populus tomentosa.Method The CDS sequence of PtoAED3 was cloned by homologous sequence from P. trichocarpa, and a prokaryotic expression vector pGEX-4T-PtoAED3 containing GST tag was constructed. The GST-PtoAED3 fusion protein was induced by IPTG and purified by GST beads. The purified protein PtoAED3 was co-incubated with the total protein extracted from P. tomentosa, then the candidate interacting proteins of PtoAED3 proteins were obtained by GST-pull down technique and analyzed by mass spectrometry technology.Result Through the identification of amino acid sequences of these proteins with mass spectrometry, a total of 128 candidate proteins interacting with PtoAED3 were screened, which involve multiple biological processes such as cell process, metabolic process, stress response, biological regulation and development process.Conclusion The GST-pull down combined mass spectrometry technology was used to screen out candidate proteins that interact with PtoAED3 in P. tomentosa, providing a preliminary direction for studying the interaction of PtoAED3 with substrates or complexes and the molecular regulatory mechanism affecting the growth and development of poplar.-
Keywords:
- PtoAED3 /
- GST-pull down /
- mass spectrometry /
- interacting protein
-
天冬氨酸蛋白酶(aspartic proteases, APs)是4大类蛋白水解酶之一,植物天冬氨酸蛋白酶(APs)都具有相似的蛋白酶特性,在酸性pH条件下APs具有蛋白水解酶的活性[1],其活性能够被Pepstatin A抑制[2]。它们具有2 个保守结构域,即天冬氨酸−苏氨酸−甘氨酸(DTG)和天冬氨酸−丝氨酸−甘氨酸(DSG),其催化活性取决于天冬氨酸残基。天冬氨酸蛋白酶(EC:3.4.23)的催化活性涉及2 个高度保守的天冬氨酸残基之间水分子的配位,APs的活性位点通常会结合在其多肽底物的6 ~ 10 个氨基酸区域,并对其进行加工[3]。因其结构特征和催化加工方式的不同,造成天冬氨酸蛋白酶功能多样化。
目前从不同植物中鉴定并纯化了多个天冬氨酸蛋白酶[4],在拟南芥(Arabidopsis thaliana)基因组、水稻(Oryza sativa)基因组和葡萄(Vitis vinifera)基因组中,分别检测到69、96、50 个编码天冬氨酸蛋白酶的基因[5-7]。植物APs 主要分为3 种类型:A类为典型的APs,B类为类珠心特异的APs,C类为非典型的APs[8-9],典型的植物APs主要含有N端的信号肽(signal peptide)、前导片段(prosegment)、ASP 结构域(ASP domain)、植物特有插入片段(plant specific insert,PSI)和C端结构域(carboxy-terminal),ASP 结构域包含2 个保守的植物ASP活性位点,即天冬氨酸−苏氨酸−甘氨酸(DTG)和天冬氨酸−丝氨酸−甘氨酸(DSG),依据是否含有PSI,可将植物典型的APs分为A1和A2两类。类珠心特异的 APs 在蛋白结构上含有N端的信号肽、ASP结构域和C端结构域,与典型植物天冬氨酸蛋白酶相比,类珠心特异的 APs不含前导片段和PSI。非典型的APs被细分为6 个亚类(C1、C2、C3、C4、C5和CX),C1亚类含有一个信号肽和一段25 ~ 30 个氨基酸残基的前导片段;C2 亚类具有转运肽特征的富丝氨酸蛋白,其中一个特征是缺乏前导片段;C3亚类包含长前导片段;C4亚类特异性具有C端延伸区段;C5亚类含有DTS活性位点而不是DTG活性位点;非典型的APs 序列中不符合上述描述的组被列入CX亚类。
天冬氨酸蛋白酶在植物生长发育和防御反应过程中起着重要作用[10]。主要参与蛋白质的储存和降解、防御反应、种子萌发、有性生殖过程,影响植物衰老和程序性死亡。例如在大麦(Hordeum vulgare)的根和叶中的天冬氨酸蛋白酶(phytepsin)定位在液泡中,参与蛋白质的加工和降解[11]。在蛋白质的储存和降解过程中,发现小麦(Triticum aestivum)WAP1和WAP2天冬氨酸蛋白酶能消化谷蛋白底物,参与了谷蛋白的降解,为种子的发育提供营养[12]。在植物衰老过程中,拟南芥CND41和烟草(Nicotiana tabacum)CND41参与了核酮糖-1,5-二磷酸羧化酶蛋白的降解,导致了叶片衰老[13-14]。在胚胎发育和有性生殖过程中,拟南芥PCS1基因异位表达时花粉囊不能正常开裂[15]。在植物防御反应过程中,拟南芥ASPG1通过ABA信号通路,介导植物干旱胁迫应答[16],而拟南芥CDR1基因表达受抑制时,植株对病原体的侵染更为敏感,说明该基因的功能与植物抵御病原体侵害相关[17]。
在木本植物中,关于天冬氨酸蛋白酶的功能研究较少,其作用机制尚未明确。Cao等人[18]在毛果杨(Populus trichocarpa)中发现了89 个编码天冬氨酸蛋白酶的基因(PtAPs),其中67 个有完整的天冬氨酸(ASP)结构域。这些PtAPs基因具有不同的组织表达模式,暗示了它们的功能多样性。其中,PtAP5、PtAP17、PtAP45和PtAP66等在木质部细胞/维管组织中特异表达,说明可能与木材形成过程有关。Bhalerao等人[19]研究发现,欧洲山杨(Populus tremula)中2 个天冬氨酸蛋白酶基因在衰老叶片中的表达量明显高于幼嫩叶片,推测其可能与衰老叶片中叶绿体的降解密切相关。
本研究通过同源克隆获得了毛白杨(Populus tomentosa) Apoplastic EDS1-dependent protein 3(PtoAED3)基因,其编码一个由425 个氨基酸组成的非典型天冬氨酸蛋白酶,其生物学功能尚未见报道,运用GST-pull down技术从毛白杨中筛选与PtoAED3相互作用的蛋白(PtoAED3 interacting proteins,PtoAED3IPs),联合质谱分析鉴定这些潜在互作蛋白的氨基酸序列并结合生物信息学功能预测,为解析毛白杨PtoAED3在植物生长发育中的分子机制奠定基础。
1. 材料与方法
1.1 试验材料和试剂
试验所用野生型毛白杨无菌组培材料幼苗,原核表达载体 pGEX-4T-1,大肠杆菌BL21(DE3)感受态均来自本实验室。通过三维结构预测获得PtoAED3的保守多肽段MQQQNHRILFDVPNSR,合成多肽免疫2只新西兰兔,共免疫6次,收集血清进行ELISA效价检测,效价 > 1∶50 000以上进行多克隆抗血清收集,血清采用柱层析方法,将特异性抗体与抗原结合,然后非特异性洗脱,透析去除离子得到特异性较好的anti-PtoAED3抗体。该抗体由北京康颂博生物科技有限公司进行合成;高保真Pfu-DNA聚合酶购自全式金生物公司;植物总RNA提取试剂盒购自艾德莱生物公司;cDNA反转录试剂盒、琼脂糖凝胶回收试剂盒与普通质粒小提试剂盒购自北京天根生物公司;限制性内切酶BamH Ⅰ和Xho Ⅰ购自NEB公司;T4-DNA连接酶购自promega公司;谷胱甘肽琼脂糖凝胶偶联物Glutathione-Sepharose购自Sigma公司;蛋白marker、G-250和核酸marker购自北京聚合美生物科技有限公司;其他化学试剂购自北京化学公司或北京国药有限公司。
1.2 PtoAED3基因的克隆及鉴定
从毛果杨数据库3.0(Phytozome)中下载PtrAED3序列,利用Premier 5设计引物,上下游引物分别为:5′-CGGGATCCCTAAACACCAGAGGCCAAGGCAC-3′,5′-CCGCTCGAGTCAAGAACATGGCTCACGAG-3′;上游引物引入BamH Ⅰ酶切位点,下游引物加入Xho Ⅰ酶切位点。按照植物总RNA提取试剂盒说明书操作提取毛白杨RNA,通过反转录试剂盒操作反转录获得cDNA作为模板,使用Pfu高保真DNA聚合酶进行PCR扩增。产物经1%琼脂糖凝胶电泳得到单一条带,按照琼脂糖凝胶回收试剂盒说明书切胶回收,将回收产物连接到pMD18-T载体,转化大肠杆菌,提取质粒后送北京华大基因公司进行DNA测序,将测序结果与GenBank中的其他物种的核苷酸序列进行序列比对。
1.3 GST-PtoAED3融合蛋白的表达和纯化
利用SignalP 4.1预测PtoAED3信号肽的序列为1 ~ 18号氨基酸。去除信号肽的PtoAED3和pGEX-4T-1空载体经过BamH Ⅰ和Xho Ⅰ双酶切后,使用T4连接酶在4 ℃过夜连接, 分别将该载体与pGEX-4T-1空载体转化大肠杆菌BL21,于20 ℃、1 mmol/L IPTG 诱导表达8 h,离心收集并裂解菌体(裂解液配方:40 mmol/L pH 7.5 Tris-HCl,150 mmol/L NaCl,1 mmol/L EDTA,0.5% NP-40,10%甘油,1 mmol/L DTT,0.4 mmol/L PMSF,2 μg/mL亮抑酶肽,2 μg/mL抑蛋白酶肽,5 mg/L溶菌酶),冰上孵育30 min,超声破碎5 min,离心取上清。pGEX-4T-1重组载体为实验组,空载体为对照,进行蛋白SDS-PAGE电泳检测蛋白表达结果。
利用谷胱甘肽琼脂糖凝胶偶联物对原核表达的GST-PtoAED3融合蛋白进行纯化去除杂蛋白,取2 mL 破碎后的GST-PtoAED3原核表达菌体上清溶液加入Glutathione-Sepharose GE柱;5 mL PBS洗柱子3次;0.5 mL洗脱液,洗脱5次;收集洗脱液并取少量的谷胱甘肽琼脂糖凝胶偶联物,用SDS-PAGE检测蛋白纯度。利用anti-PtoAED3抗体对纯化后的GST-PtoAED3蛋白进行Western杂交检测。
1.4 GST-pull down筛选PtoAED3的互作蛋白
选取3月龄的野生型毛白杨组培苗植株,加液氮研磨,加入PBS缓冲液提取植株总蛋白。将毛白杨总蛋白分别与带有GST或GST-PtoAED3蛋白的谷胱甘肽琼脂糖凝胶偶联物共孵育3 h(融合蛋白已亲和固化在Glutathione-Sepharose beads上),然后用蛋白提取缓冲液对共孵育过的Glutathione-Sepharose beads进行清洗。最后用高浓度的还原性谷胱甘肽溶液进行洗脱,收集蛋白溶液,用于后续分析,其中GST-PtoAED3组为实验组,GST组为对照组。利用SDS-PAGE电泳检测洗脱下来的蛋白样品,并用0.25%考马斯亮蓝G-250染色3 h。
1.5 LC-MS/MS分析
向pull down得到的PtoAED3IPs与GSTIPs中加入胰蛋白酶对蛋白质样品进行酶解,然后使用LC-MS/MS(nanoLC-QE)进行质谱分析,最后使用MASCOT等质谱匹配软件分析LC-MS/MS数据,获得与PtoAED3相结合的候选蛋白质的信息。通过上海中科新生命有限公司的Mascot软件在最新的NCBI数据库(http://www.ncbi.nlm.nih.gov,National Center for Biotechnology Information)和 UniProtKB /Swiss-Prot数据库(http://www.Uniprot.org,UniProt Knowledgebase)中对PtoAED3IPs蛋白进行肽段指纹图谱蛋白的检索。
1.6 生物信息学分析
从检索的PtoAED3IPs蛋白和GSTIPs蛋白中挑选差异化的蛋白,并进行序列比对(Blast),随后批量提取目标蛋白及其Blast hits相关的GO term(Mapping),并进行GO Annotation,最后通过InterProScan和ANNEX进行补充注释。
2. 结果与分析
2.1 毛白杨PtoAED3基因的克隆
利用RNA提取试剂盒获得质量较好且未降解的毛白杨总RNA(图1A)。通过cDNA反转录试剂盒获得毛白杨cDNA,作为克隆PtoAED3的模板,PCR扩增后将产物进行琼脂糖凝胶电泳观察到约1 200 bp处1条明亮的特异性条带(图1B),通过基因测序并与毛果杨PtrAED3比对后,确定得到PtoAED3基因长度为1 278 bp(图1C),含有3 个外显子,2 个内含子。其蛋白序列含425 个氨基酸,活性位点为DSG/DTG,属于非典型类型的天冬氨酸蛋白酶(图1C)。通过信号肽SignalP4.1预测表明,毛白杨PtoAED3蛋白在N端存在18 个氨基酸长度的信号肽(图1D)。
![]() 图 1 毛白杨PtoAED3
图 1 毛白杨PtoAED3基因的克隆 A. 毛白杨的总RNA;B. 毛白杨PtoAED3 PCR结果;C. 毛白杨PtoAED3核酸和PtoAED3蛋白序列信息;D. 毛白杨 PtoAED3 信号肽预测。A, total RNA extraction result of Populus tomentosa; B, PCR result of PtoAED3 cDNA in Populus tomentosa; C, sequence information of PtoAED3 nucleic acid and PtoAED3 protein; D, signal peptide prediction of PtoAED3 protein.Figure 1. Results of PtoAED3 gene cloning in P. tomentosa2.2 GST-PtoAED3融合蛋白的原核表达和纯化
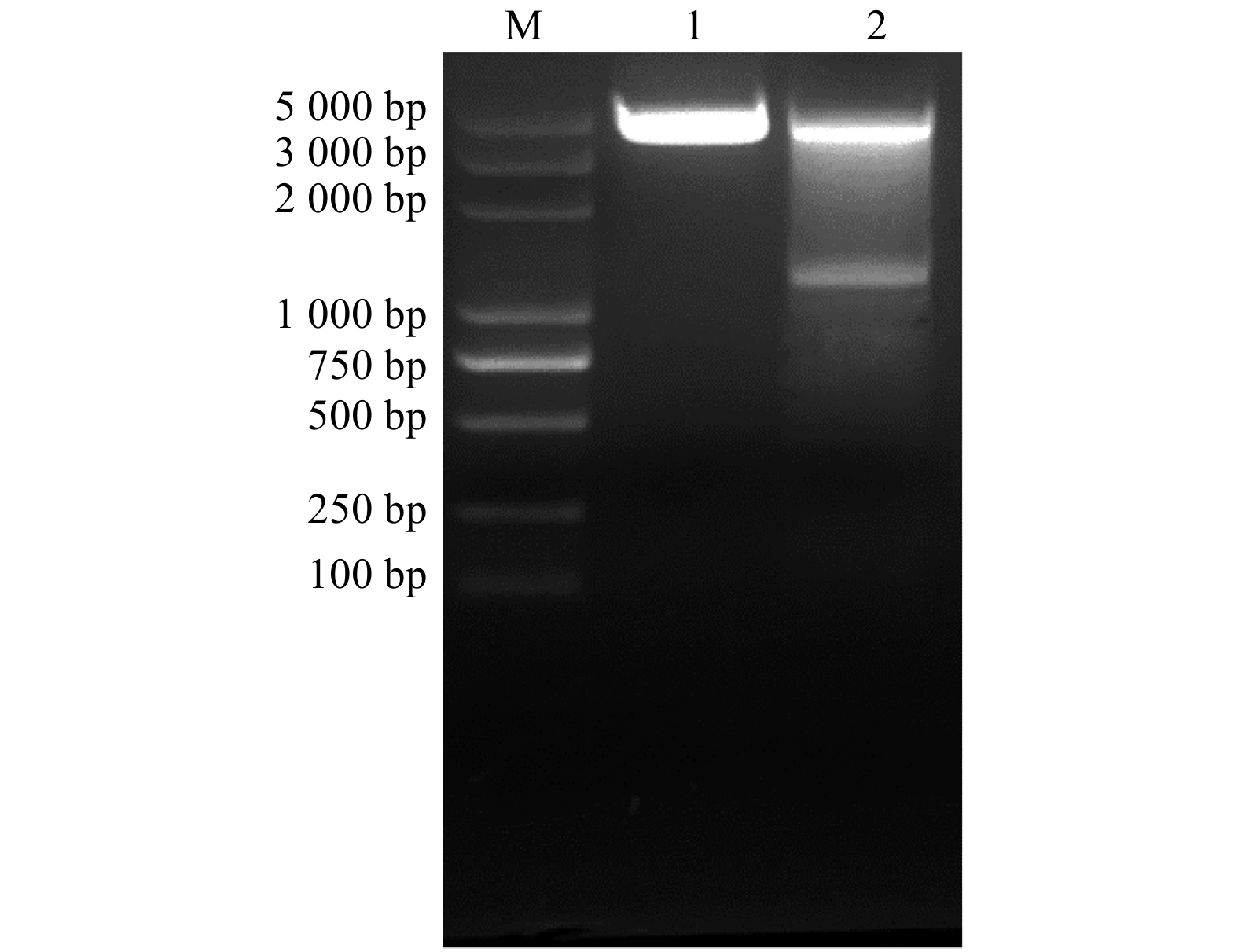
为了表达带有GST标签的GST-PtoAED3融合蛋白,克隆获得去除信号肽的PtoAED3,与BamH Ⅰ/Xho Ⅰ双酶切后的pGEX-4T-1载体进行连接,获得pGEX-4T-PtoAED3重组载体,双酶切后电泳分析,插入的目的片段与预期PtoAED3大小(1 224 bp)一致(图2)。
![]() 图 2 pGEX-4T-PtoAED3原核表达载体双酶切电泳图M. 2 000 bp DNA Marker;泳道1. pGEX-4T-1空载体双酶切电泳图;泳道2. pGEX-4T-PtoAED3双酶切电泳图。Lane 1, electrophoresis result of pGEX-4T-1 empty vector after BamHⅠ/XhoⅠ digestion; lane 2, electrophoresis result of pGEX-4T-PtoAED3 after BamHⅠ/XhoⅠ digestion.Figure 2. Double enzyme digestion of pGEX-4T-PtoAED3 vector from P. tomentosa
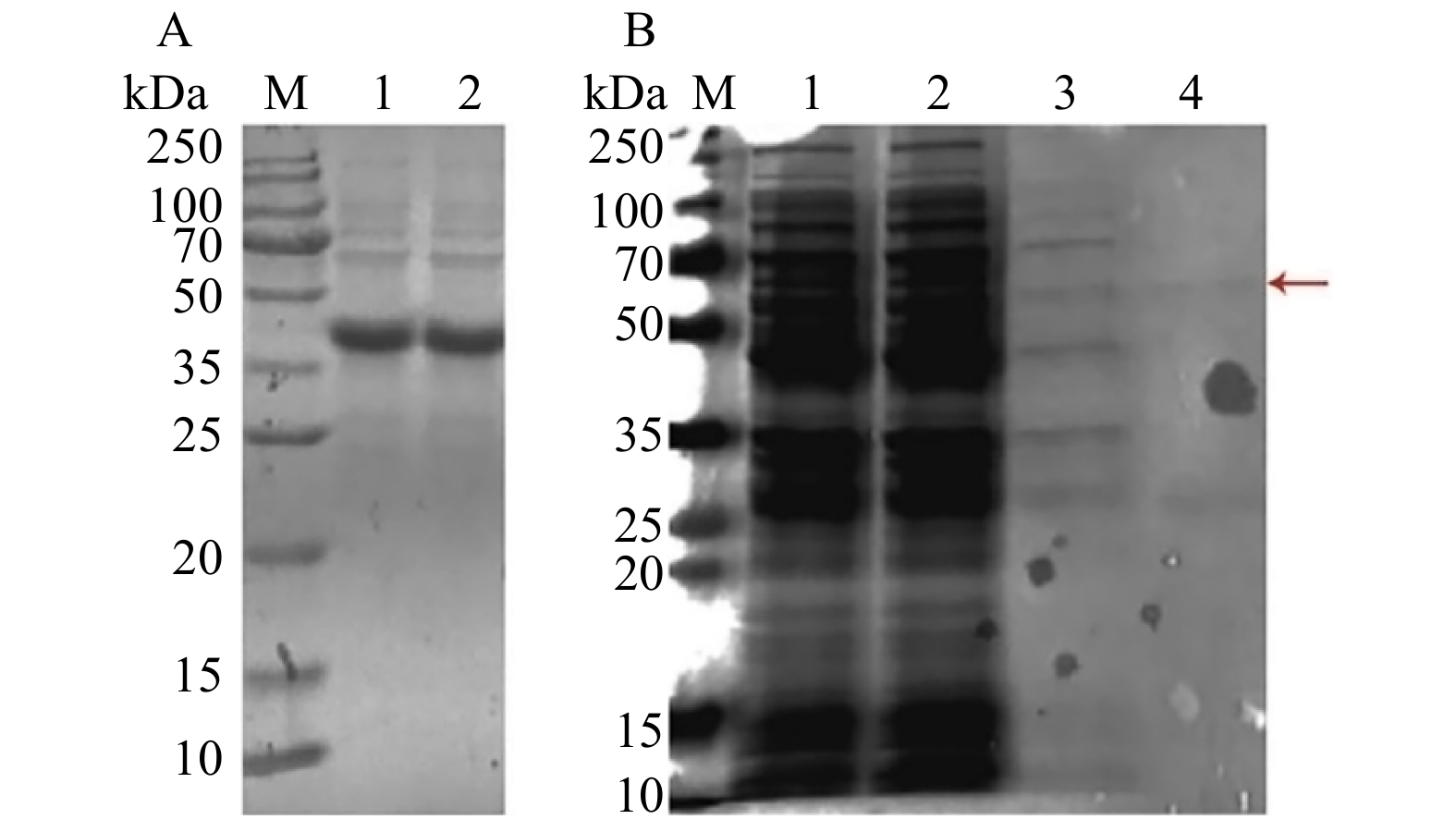
图 2 pGEX-4T-PtoAED3原核表达载体双酶切电泳图M. 2 000 bp DNA Marker;泳道1. pGEX-4T-1空载体双酶切电泳图;泳道2. pGEX-4T-PtoAED3双酶切电泳图。Lane 1, electrophoresis result of pGEX-4T-1 empty vector after BamHⅠ/XhoⅠ digestion; lane 2, electrophoresis result of pGEX-4T-PtoAED3 after BamHⅠ/XhoⅠ digestion.Figure 2. Double enzyme digestion of pGEX-4T-PtoAED3 vector from P. tomentosa将携带pGEX-4T-1、pGEX-4T-PtoAED3质粒的BL21大肠杆菌采用37 ℃、1 mmol/L IPTG诱导表达3 h,沉淀中含有大量的包涵体蛋白,上清液中未检测到目的蛋白。因此通过在20 ℃、1 mmol/L IPTG诱导表达8 h,在上清液中检测到大量表达的PtoAED3蛋白,大小为65 kDa,用于后续蛋白纯化实验(图3A)。将表达GST、GST-PtoAED3融合蛋白的大肠杆菌进行超声裂解提取蛋白,将全菌蛋白加入谷胱甘肽(GST)琼脂糖凝胶偶联物中,利用含有GST标签的融合蛋白能结合谷胱甘肽琼脂糖凝胶偶联物的原理,得到纯化的GST-PtoAED3融合蛋白,在65 kDa大小位置存在单一的目的条带(图3B)。将纯化后的GST-PtoAED3蛋白与抗体anti-PtoAED3(MQQQNHRILFDVPNSR)进行免疫杂交检测,目的条带单一(图3C),可用于后续pull-down实验。
![]() 图 3 毛白杨PtoAED3基因表达和纯化A. 毛白杨PtoAED3基因原核表达SDS-PAGE电泳图;泳道1. IPTG未诱导的pGEX-4T-1上清溶液; 泳道2. IPTG未诱导的pGEX-4T-PtoAED3上清溶液; 泳道3. IPTG诱导的pGEX-4T-1蛋白表达上清溶液; 泳道4.IPTG诱导的pGEX-4T-PtoAED3蛋白表达上清溶液; 泳道5. IPTG诱导的pGEX-4T-1菌体沉淀; 泳道6. IPTG诱导的pGEX-4T-PtoAED3菌体沉淀。B. 利用GST标签蛋白纯化GST-PtoAED3蛋白的SDS-PAGE电泳图;泳道1 ~ 3分别为洗脱5、3、1次后,谷胱甘肽琼脂糖凝胶偶联物通过SDS-PAGE检测。C. 抗体anti-PtoAED3与纯化后的原核表达蛋白免疫杂交;泳道1. 抗体与IPTG诱导的pGEX-4T-1原核表达蛋白杂交; 泳道2. 抗体与IPTG诱导的pGEX-4T-PtoAED3原核表达蛋白杂交。红色箭头表示原核表达的目的蛋白。A, SDS-PAGE electrophoresis of PtoAED3 prokaryotic expression; lane 1, non-IPTG induced pGEX-4T-1 protein expression in supernatant; lane 2, non-IPTG induced pGEX-4T-PtoAED3 protein expression in supernatant; lane 3, IPTG induced pGEX-4T-1 protein expression in supernatant; lane 4, IPTG induced pGEX-4T-PtoAED3 protein expression in supernatant; lane 5, IPTG induced pGEX-4T-1 protein expression in precipitation; lane 6, IPTG induced pGEX-4T-PtoAED3 protein expression in precipitation. B, SDS-PAGE electrophoresis results using GST-tagged protein purification for GST-PtoAED3 protein; lane 1−3, the glutathione-sepharose eluted 5,3,1 times are detected by SDS-PAGE. The red arrow indicates the interested proteins expressed in E.coli. C, western blotting of anti-PtoAED3 antibody with the purified prokaryotic expression protein; lane 1, western blotting of anti-PtoAED3 antibody with the IPTG-induced pGEX-4T-1 prokaryotic expression protein; lane 2, western blotting of anti-PtoAED3 antibody with the IPTG pGEX-4T-PtoAED3 protein hybridization. The red arrow indicates the protein of interest expressed prokaryotically.Figure 3. Expression and purification of PtoAED3 in P. tomentosa
图 3 毛白杨PtoAED3基因表达和纯化A. 毛白杨PtoAED3基因原核表达SDS-PAGE电泳图;泳道1. IPTG未诱导的pGEX-4T-1上清溶液; 泳道2. IPTG未诱导的pGEX-4T-PtoAED3上清溶液; 泳道3. IPTG诱导的pGEX-4T-1蛋白表达上清溶液; 泳道4.IPTG诱导的pGEX-4T-PtoAED3蛋白表达上清溶液; 泳道5. IPTG诱导的pGEX-4T-1菌体沉淀; 泳道6. IPTG诱导的pGEX-4T-PtoAED3菌体沉淀。B. 利用GST标签蛋白纯化GST-PtoAED3蛋白的SDS-PAGE电泳图;泳道1 ~ 3分别为洗脱5、3、1次后,谷胱甘肽琼脂糖凝胶偶联物通过SDS-PAGE检测。C. 抗体anti-PtoAED3与纯化后的原核表达蛋白免疫杂交;泳道1. 抗体与IPTG诱导的pGEX-4T-1原核表达蛋白杂交; 泳道2. 抗体与IPTG诱导的pGEX-4T-PtoAED3原核表达蛋白杂交。红色箭头表示原核表达的目的蛋白。A, SDS-PAGE electrophoresis of PtoAED3 prokaryotic expression; lane 1, non-IPTG induced pGEX-4T-1 protein expression in supernatant; lane 2, non-IPTG induced pGEX-4T-PtoAED3 protein expression in supernatant; lane 3, IPTG induced pGEX-4T-1 protein expression in supernatant; lane 4, IPTG induced pGEX-4T-PtoAED3 protein expression in supernatant; lane 5, IPTG induced pGEX-4T-1 protein expression in precipitation; lane 6, IPTG induced pGEX-4T-PtoAED3 protein expression in precipitation. B, SDS-PAGE electrophoresis results using GST-tagged protein purification for GST-PtoAED3 protein; lane 1−3, the glutathione-sepharose eluted 5,3,1 times are detected by SDS-PAGE. The red arrow indicates the interested proteins expressed in E.coli. C, western blotting of anti-PtoAED3 antibody with the purified prokaryotic expression protein; lane 1, western blotting of anti-PtoAED3 antibody with the IPTG-induced pGEX-4T-1 prokaryotic expression protein; lane 2, western blotting of anti-PtoAED3 antibody with the IPTG pGEX-4T-PtoAED3 protein hybridization. The red arrow indicates the protein of interest expressed prokaryotically.Figure 3. Expression and purification of PtoAED3 in P. tomentosa2.3 GST-pull down联合质谱筛选和鉴定毛白杨PtoAED3的互作蛋白
为了筛选与毛白杨PtoAED3相互作用的蛋白,将多株毛白杨组培苗混合样品提取的全蛋白(图4A),分别与结合了GST或GST-PtoAED3的Glutathione-Sepharose beads共孵育3 h,然后利用还原型谷胱甘肽对孵育的全蛋白进行洗脱,当洗脱5 次时,SDS-PAGE电泳条带仅含GST、GST-PtoAED3蛋白条带(图4B、泳道4),利用LC-MS/MS联合质谱仪对GST-pull down洗脱下来的PtoAED3IPs蛋白和GSTIPs蛋白进行质谱检测。
![]() 图 4 利用GST-pull down在毛白杨中筛选与GST-PtoAED3互作的蛋白A. 毛白杨植株全蛋白电泳图; B. 还原型谷胱甘肽洗脱与GST-PtoAED3互作的蛋白。泳道1和2为未洗脱的植株全蛋白,泳道3和4分别为利用还原型谷胱甘肽洗脱3和5次收集的蛋白。A, electrophoresis of whole protein of P. tomentosa; B, GST-PtoAED3 interacting protein eluted with reduced glutathione. Lane 1 and 2, uninteracting proteins; lane 3 and 4, proteins with reduced glutathione detected by SDS-PAGE for 3 and 5 times.Figure 4. Screening of proteins interacting with GST-PtoAED3 in P. tomentosa by GST-pull down
图 4 利用GST-pull down在毛白杨中筛选与GST-PtoAED3互作的蛋白A. 毛白杨植株全蛋白电泳图; B. 还原型谷胱甘肽洗脱与GST-PtoAED3互作的蛋白。泳道1和2为未洗脱的植株全蛋白,泳道3和4分别为利用还原型谷胱甘肽洗脱3和5次收集的蛋白。A, electrophoresis of whole protein of P. tomentosa; B, GST-PtoAED3 interacting protein eluted with reduced glutathione. Lane 1 and 2, uninteracting proteins; lane 3 and 4, proteins with reduced glutathione detected by SDS-PAGE for 3 and 5 times.Figure 4. Screening of proteins interacting with GST-PtoAED3 in P. tomentosa by GST-pull down质谱结果表明,实验组中共检测到195 个与GST-PtoAED3互作的蛋白,对照组中共检测到158 个与GST互作的蛋白。将对照组与实验组中都检测到的蛋白从实验组数据中删除,去除背景,得到128 个可能与PtoAED3互作的候选蛋白。
2.4 毛白杨PtoAED3的互作蛋白功能分析
根据质谱,得到肽段评分大于20分的与PtoAED3相互作用的候选蛋白有128 个,其中23 个为未知的蛋白,105 个为已知蛋白,对105 个已知蛋白进行GO功能分类分析。Level 2级别GO功能富集词条总计43 个,其中生物学过程21 个,占总数48.8%,分子功能7 个,占总数16.3%,细胞组成部分15 个,占总数34.9%(图5)。
表 1 GST-pull down筛选的PtoAED3互作蛋白Table 1. PtoAED3 interacting proteins screened by GST-pull down项目
Item蛋白编号
Protein No.肽段
Peptide segment蛋白注释
Protein annotation肽段得分
Peptide score覆盖率
Coverage/%高丰度蛋白
High abundance
proteinQ8LPG9 K.EDVEALWLK.K/
R.SQAIIK.LProtein IQ-DOMAIN 14
蛋白IQ结构域20.84/21.91 3.31 Q56XU4 R.MKM*DR.E Zinc finger CCCH domain-containing protein 6
锌指结构域26.92 1.15 C6E2H7 K.KLEWYEK.F GTPase Era
GTP 水解酶27.89 1.99 F4I0N3 R.ELQQLK.E WEB family protein
WEB 转录因子39.89 2.91 O04590 R.VAAEALIR.L Pentatricopeptide repeat-containing protein
三角状五肽23.91 4.88 蛋白质加工降解
Protein processing
and degradationQ9D832 K.NPVNK.K DnaJ homolog subfamily B member 4
分子伴侣21.05 1.58 P43297 K.EGAVAEVK.D Cysteine proteinase RD21A
半胱氨酸蛋白酶20.47 1.72 Q9C5D2 R.MQENISR.F F-box/LRR-repeat protein 4
SCF复合体亚基25.39 1.94 Q9FZK1 K.ILGGKATWATTK.C F-box only protein 6
SCF 复合体亚基21.78 3.22 Q94AH6 R.LLFDK.S Cullin-1 SCF 复合体亚基 37.5 1.22 Q94F30 K.FTLSR.V Ubiquitin-like-specific protease ESD4
泛素特异蛋白酶22.41 0.97 Q9JIG7 R.QNLGKAK.L Coiled-coil domain-containing protein 22
螺旋卷曲结构域蛋白40.1 1.67 O22993 R.FTLSR.S Probable inactive ATP-dependent zinc
metalloprotease FTSHI 1
金属蛋白酶22.41 0.62 胁迫响应相关
Stress response
correlationP43297 K.EGAVAEVK.D Cysteine proteinase RD21A
半胱氨酸蛋白酶20.47 1.72 O80396 M.AGLEELK.K Mitogen-activated protein kinase kinase 3
分裂原活化蛋白激酶27.02 1.33 O04203 R.LPELR.K Nematode resistance protein-like HSPRO2
类线虫抗性蛋白24.23 5.38 Q9LII8 R.IDSELR.H Protein KINESIN LIGHT CHAIN-RELATED 2
驱动蛋白轻链30.55 3.61 O81825 K.LTELR.K Probable disease resistance protein At4g27220
抗病蛋白26.75 0.83 Q9LFP7 R.NVGLK.T Probable serine/threonine-protein kinase PIX7
丝氨酸/苏氨酸蛋白激酶21.76 1.13 Q7XA40 R.SLDELK.R Putative disease resistance protein RGA3
抗病蛋白28.92 7.69 Q9LRR4 R.LLEIR.A Putative disease resistance RPP13-like protein 1
抗病蛋白28.42 1.79 Q9LX93 K.LPRSR.M E3 ubiquitin-protein ligase RING1
E3 泛素连接酶22.21 17.86 Q7VG78 K.AGPLSARKPASK.Q Probable GMP synthase [glutamine-hydrolyzing]
GMP合成酶22.29 23.53 Q9C5D2 R.MQENISR.F F-box/LRR-repeat protein 4
SCF 复合体亚基25.39 1.94 Q0WQF4 K.ITELR.E Vacuolar protein sorting-associated protein 53 A
液泡蛋白分类相关蛋白26.75 1.48 O22769 R.LLEIR.Q NADH dehydrogenase [ubiquinone] flavoprotein 2
NADH 脱氢酶[泛醌]黄素蛋白28.42 1.97 Q9UI10 K.IIIADK.V Translation initiation factor eIF-2B subunit delta
翻译起始因子亚基25.12 0.94 O65202 R.ILELR.L Peroxisomal acyl-coenzyme A oxidase 1
过氧化物酶体酰基辅酶A氧化酶28.42 0.75 F4I9E1 R.LKM*IPR.T Protein NUCLEAR FUSION DEFECTIVE 4
核融合缺陷蛋白25.57 1.06 P47140 R.EQSNVIAR.Q Altered inheritance rate of mitochondria protein 25
线粒体蛋白21.26 2.31 O43099 R.LPELR.K Peroxiredoxin Asp f3
过氧化物酶24.23 4.13 Q9FYA2 K.ENYHLVPR.T Probable WRKY transcription factor 75
WRKY转录因子32.11 6.35 Q84X53 R.SLDELK.E Transcription termination factor MTEF1, chloroplastic
转录终止因子28.92 2.48 发育过程相关
Developmental
processQ9UI10 K.IIIADK.V Translation initiation factor eIF-2B subunit delta
翻译起始因子亚基25.12 0.94 Q84JG2 K.ILRFEELDLQM*EK.E SWI/SNF complex subunit SWI3B
SWI/SNF复合体亚基23.07 2.60 Q12955 K.AGMTPLDLATNEEIR.L Ankyrin-3 锚蛋白 24.76 8.72 Q8GWQ6 K.AYALDK.K UPF0235 protein At5g63440 UPF蛋白 21.38 6.25 Q84X53 R.SLDELK.E Transcription termination factor MTEF1, chloroplastic
转录终止因子28.92 2.48 F4JRR5 R.VDTFR.I TITAN-like protein
TITAN蛋白21.33 3.03 Q94AH6 R.LLFDK.S Cullin-1
SCF 复合体亚基37.5 1.22 O22993 R.FTLSR.S Probable inactive ATP-dependent zinc metalloprotease FTSHI 1
金属蛋白酶22.41 0.62 Q9LII8 R.IDSELR.H Protein KINESIN LIGHT CHAIN-RELATED 2
驱动蛋白轻链30.55 3.61 Q0D4T4 K.VELKR.I MADS-box transcription factor 18
MADS转录因子23.86 2.53 Q9ZVP8 K.VLEQR.A NAC domain-containing protein 35
NAC转录因子25.43 1.98 Q94F30 K.FTLSR.V Ubiquitin-like-specific protease ESD4
泛素特异蛋白酶22.41 0.97 Q9FYA2 K.ENYHLVPR.T Probable WRKY transcription factor 75
WRKY转录因子32.11 6.35 Q9FII5 R.LGGMIPAK.T Leucine-rich repeat receptor-like protein kinase TDR
受体蛋白激酶23.78 5.88 Q9LHP4 R.IPELR.T Receptor-like protein kinase 2
受体蛋白激酶24.23 3.57 Q9SN39 R.ILFDK.Y Pentatricopeptide repeat-containing protein DOT4
三角状五肽蛋白37.5 2.53 F4I9E1 R.LKM*IPR.T Protein NUCLEAR FUSION DEFECTIVE 4
核融合缺陷蛋白25.57 1.06 Q8LEZ4 R.AGVERK.A Protein NUCLEAR FUSION DEFECTIVE 5
核融合缺陷蛋白25.82 0.84 Q9FZK1 K.ILGGKATWATTK.C F-box only protein 6
SCF 复合体亚基21.78 3.22 Q9M811 K.EAPQVPASPR.E Rop guanine nucleotide exchange factor 11
鸟苷酸交换因子21.47 7.41 Q2QDF6 .LSEVR.G Steroid 5-alpha-reductase DET2
类固醇还原酶22.75 6.33 ![]() 图 5 PtoAED3互作蛋白GO功能富集Cellular processes 生物学途径包括细胞进程; Metabolism process 代谢过程; Response to stimulus 应激反应; Biological regulation 生物调节; Multicellular organismal process 多细胞器官进程; Regulation of biological process 生物进程调节; Development process 发育过程; Cellular component organization or biogenesis 细胞组分组成或发育; Localization 定位; Reproductive process 繁殖过程; Positive regulation of biological process 生物过程正调节; Multi-organism process 多器官进程; Signaling 信号途径; Negative regulation of biological process 生物过程负调节; Rhythmic process 节律进程; Growth 生长; Immune system process 免疫体统进程; Pigmentation 色素沉积; Biological adhesion 生物固定; Carbon utilization 碳利用; Binding 分子功能包括结合; Catalytic activity 催化活性; Transporter activity 转运活性; Transcription regulator activity 转录调节因子活性; Structural molecular activity 结构分子活性; Antioxidant activity 抗氧化活性; Cell part 细胞组分则包括细胞部分; Cell 细胞; Organelle 细胞器; Membrane 膜; Membrane part 膜组分; Organelle part 细胞器组分; Protein-containing complex 蛋白复合体; Membrane-enclosed lumen 膜封闭腔; Cell junction 细胞连接; Symplast 共质体; Extracellular region 胞外域; Supramolecular complex 超分子复合体; Synapse 突触; Synaptic part and extracellular region part 突触局部及胞外域局部Figure 5. GO function enrichment of PtoAED3IPs
图 5 PtoAED3互作蛋白GO功能富集Cellular processes 生物学途径包括细胞进程; Metabolism process 代谢过程; Response to stimulus 应激反应; Biological regulation 生物调节; Multicellular organismal process 多细胞器官进程; Regulation of biological process 生物进程调节; Development process 发育过程; Cellular component organization or biogenesis 细胞组分组成或发育; Localization 定位; Reproductive process 繁殖过程; Positive regulation of biological process 生物过程正调节; Multi-organism process 多器官进程; Signaling 信号途径; Negative regulation of biological process 生物过程负调节; Rhythmic process 节律进程; Growth 生长; Immune system process 免疫体统进程; Pigmentation 色素沉积; Biological adhesion 生物固定; Carbon utilization 碳利用; Binding 分子功能包括结合; Catalytic activity 催化活性; Transporter activity 转运活性; Transcription regulator activity 转录调节因子活性; Structural molecular activity 结构分子活性; Antioxidant activity 抗氧化活性; Cell part 细胞组分则包括细胞部分; Cell 细胞; Organelle 细胞器; Membrane 膜; Membrane part 膜组分; Organelle part 细胞器组分; Protein-containing complex 蛋白复合体; Membrane-enclosed lumen 膜封闭腔; Cell junction 细胞连接; Symplast 共质体; Extracellular region 胞外域; Supramolecular complex 超分子复合体; Synapse 突触; Synaptic part and extracellular region part 突触局部及胞外域局部Figure 5. GO function enrichment of PtoAED3IPs根据PepCount指数即质谱过程中每个蛋白检测到的肽段数对所有pull-down结果蛋白进行排序,得到5 个检测丰度较高的蛋白(表1)。Uniport数据库中的功能注释表明,蛋白IQ结构域Protein IQ-DOMAIN 14涉及与钙调蛋白或钙调蛋白样蛋白的相互作用,也参与转录或转录后水平调控基因表达。GTPase Era是一种结合GDP与GTP并快速交换核苷酸所必需的GTP水解酶,参与16S rRNA加工和30S核糖体亚基生物合成,还参与细胞周期调控和能量代谢。转录因子Zinc finger CCCH domain-containing protein 6则涉及金属离子结合、DNA结合。转录因子WEB family protein涉及叶绿体运动相关功能。三角状五肽Pentatricopeptide repeat-containing protein涉及RNA修饰,胞嘧啶到尿嘧啶的修饰等功能。
筛选到1 个与蛋白质折叠(protein folding)相关和7 个与蛋白质降解(proteolysis)相关的蛋白(表1)。DnaJ homolog subfamily B member 4作为蛋白分子伴侣,在动物研究中发现,它能在体外促进ATP水解和HSPA1A/B介导的未折叠蛋白的折叠[20]。Coiled-coil domain-containing protein 22参与NF-kappa-B信号通路的调控,促进I-kappa-B激酶IKBKB亚基的泛素化及随后的蛋白酶体降解,该功能可能是通过与COMMD1相关并涉及cul2依赖性E3泛素连接酶复合物下调NF-kappa-B活性而实现的[21]。在拟南芥中,Ubiquitin-like-specific protease ESD4加工SUMO前体生成成熟SUMO,与野生型相比,esd4突变体含有较少的游离SUMO和更多的SUMO偶联物,说明ESD4在植物体内起到从偶联物中回收SUMO的作用[22]。
筛选到20 个与防御反应(defense response)和胁迫响应(response to stress)相关的蛋白(表1)。其中半胱氨酸蛋白酶RD21A在免疫、衰老、生物与非生物胁迫中发挥作用,它参与了对坏死真菌病原体Sclerotina sclerotiorum的免疫[23],当拟南芥感染病菌时,RD21A促进了PCD进程[24],还可能参与应激或损伤细胞的死亡[25]等过程。对Mitigen-activated protein kinase kinase 3的研究发现,转基因ProMKK3: GUS株系在紫丁香假单胞菌番茄株(Pseudomonas syringaepv. tomato)DC3000强诱导下的维管组织中表达,在mkk3敲除植株中毒性Pst DC3000生长增加,而在MKK3过表达植株中毒性Pst DC3000生长减少,说明MKK3在病原菌防御中起重要作用[26]。在被P. syringae pv. tomato 侵染后的野生型拟南芥中,Nematode resistance protein-like HSPRO2表达上调,且HSPRO2突变体对P. syringae pv. tomato更敏感,说明HSPRO2是基础抗性的阳性调节因子,并在植物基础抗性中起到重要作用[27]。E3 ubiquitin-protein ligase RING1通过促进PCD负调控因子的降解参与程序性细胞死亡(PCD)的正调控,从而参与了植物防御信号通路的早期步骤 [28]。拟南芥AtVPS53(HIT1)可以与AtVPS52、AtVPS54相互结合,形成一个复合体,在热应激条件下hit1-1突变体的细胞膜稳定性低于野生型,说明AtVPS53参与植物细胞质膜的热适应,以响应热胁迫[29-30]。烟曲霉(Aspergillus fumigatus)的过氧化物酶Asp f3可以灭活ROS,asp f3缺失突变体对ROS敏感,说明Asp f3参与了细胞对氧化应激的反应[31]。SOLDAT10编码与人类线粒体转录终止因子mTERF相关的蛋白,在flu突变体基础上进行flu/soldat10双突,使flu突变体中单线氧介导的表型得以回复,说明mTERF与FLU呈拮抗关系,在拟南芥中共同响应单线氧胁迫[32]。
筛选到21个与发育过程(developmental process)相关的蛋白(表1)。对拟南芥SWI3B的研究发现,AtSWI3蛋白参与SWI/SNF复合物的组装,影响胚胎发生[33]。TITAN-like蛋白TTL是胚乳核分裂的关键调控因子,纯合突变导致拟南芥胚胎致死[34]。SCF复合体中的Cullin-1蛋白在拟南芥胚胎发育中发挥关键作用,该基因突变后造成早期胚胎发育受阻[35],Cullin-1的另一个拟南芥突变体axr6表现出根和下胚轴发育缺陷的表型,说明Cullin-1通过影响植物对生长素的反应,影响植株的早期发育[36]。SCF复合体中的F-box only protein 6(LEAF CURLING RESPONSIVENESS, LCR)可以通过影响拟南芥对生长素的响应来影响叶片的发育,其lcr突变体表现出半矮化、叶形改变和茎短等多效性缺陷[37]。金属蛋白酶FTSHI 1定位于拟南芥叶绿体包膜,参与叶绿体的形成,arc1(ftsHi1-1)突变体干扰幼苗类囊体发育,T-DNA插入突变体ftsHi1-2导致胚胎死亡[38]。转录因子MADS-box 18参与花与腋芽的发育,在水稻中过表达OsMADS18导致早花及腋芽分生组织形成[39]。转录因子NAC domain-containing protein 35作为开花抑制因子,通过影响拟南芥开花时间和冷响应两条交叉通路来共同调节花发育[40]。泛素特异蛋白酶ESD4是能将SUMO前体加工为成熟SUMO的蛋白酶,在调控拟南芥花期中发挥重要作用[22]。转录因子WRKY75调节根的发育,在Pi缺失过程中被诱导,当WRKY75表达被抑制时,拟南芥侧根长度和根毛数量显著增加[41]。Rop鸟苷酸交换因子RopGEF11是光敏色素介导的根初级发育的负调控因子,通过依赖光敏色素的方式激活细胞质中的ROPs来调节根的发育,拟南芥的RopGEF11功能缺失突变体pirf1在黑暗条件下根过度生长,而过表达突变体(PIRF1- ox)根系生长延迟且根毛不规则[42]。因此,我们发现与PtoAED3相互作用的蛋白主要集中在与胁迫相关和发育相关的生物学功能中,可能会在毛白杨抗病、耐热、抗氧化及胚胎发育、生长素响应、叶绿体发育、花发育等过程中发挥作用。128个候选蛋白中尚未提及的部分,由于篇幅原因另文叙述。
3. 讨论与结论
通过同源克隆获得了毛白杨PtoAED3基因,与其他物种AED3进行氨基酸序列比对发现AED3的两个保守天冬氨酸催化位点分别为DTS与DSG,根据Faro and Gal 2005分类标准,“C5亚类非典型天冬氨酸蛋白酶的第一个催化位点为DTS,而不是DTG”可以确定,PtoAED3为C5亚类非典型天冬氨酸蛋白酶。该类天冬氨酸蛋白酶中,已报导的拟南芥组成型抗病性基因(CDR1)编码一种质外体非典型天冬氨酸蛋白酶,它的过表达造成了植株矮化和对丁香假单胞菌抗性的增强,并通过水杨酸依赖的微氧化应激和各种防御相关基因的激活,参与了抗病性信号传导[17]。
目前,毛白杨天冬氨酸蛋白酶基因PtoAED3的功能与分子机制未知。蛋白酶通常会与其他蛋白相互作用形成复合物来发挥其生物学功能,本研究同源克隆了PtoAED3并在体外原核表达了GST-PtoAED3融合蛋白,通过GST-pull down与质谱技术鉴定了毛白杨中与PtoAED3互作的蛋白,通过对128 个可能与PtoAED3互作的蛋白进行GO功能分析,发现这些互作蛋白涉及细胞进程、代谢过程、应激反应、生物调节、发育过程等多种生物进程,说明PtoAED3可能参与前体蛋白的加工,蛋白质的降解,从而调控细胞程序性死亡(PCD),在植物抗病、抗逆及生长发育等过程中发挥重要作用。因此探寻毛白杨植株内与PtoAED3互作的蛋白可为后续PtoAED3生物学功能与分子机制的研究奠定基础。
体内蛋白互作比体外互作的关系更错综复杂,所以体外互作蛋白实验与体内真实互作存在一定差异性,因此Pull down筛选互作蛋白存在假阳性。这些筛选出的候选互作蛋白是否在体内与PtoAED3相互作用,后续需要通过BiFC等实验进行验证,而且这些蛋白是否在杨树中与PtoAED3一起参与调控生长发育,或者参与抗病抗逆相关进程,有待进一步实验验证。综上所述,本研究的结果为进一步探寻毛白杨天冬氨酸蛋白酶基因PtoAED3分子机制和生物学功能提供了线索,为后续研究提供了方向。
-
图 1 毛白杨PtoAED3
基因的克隆 A. 毛白杨的总RNA;B. 毛白杨PtoAED3 PCR结果;C. 毛白杨PtoAED3核酸和PtoAED3蛋白序列信息;D. 毛白杨 PtoAED3 信号肽预测。A, total RNA extraction result of Populus tomentosa; B, PCR result of PtoAED3 cDNA in Populus tomentosa; C, sequence information of PtoAED3 nucleic acid and PtoAED3 protein; D, signal peptide prediction of PtoAED3 protein.
Figure 1. Results of PtoAED3 gene cloning in P. tomentosa
图 2 pGEX-4T-PtoAED3原核表达载体双酶切电泳图
M. 2 000 bp DNA Marker;泳道1. pGEX-4T-1空载体双酶切电泳图;泳道2. pGEX-4T-PtoAED3双酶切电泳图。Lane 1, electrophoresis result of pGEX-4T-1 empty vector after BamHⅠ/XhoⅠ digestion; lane 2, electrophoresis result of pGEX-4T-PtoAED3 after BamHⅠ/XhoⅠ digestion.
Figure 2. Double enzyme digestion of pGEX-4T-PtoAED3 vector from P. tomentosa
图 3 毛白杨PtoAED3基因表达和纯化
A. 毛白杨PtoAED3基因原核表达SDS-PAGE电泳图;泳道1. IPTG未诱导的pGEX-4T-1上清溶液; 泳道2. IPTG未诱导的pGEX-4T-PtoAED3上清溶液; 泳道3. IPTG诱导的pGEX-4T-1蛋白表达上清溶液; 泳道4.IPTG诱导的pGEX-4T-PtoAED3蛋白表达上清溶液; 泳道5. IPTG诱导的pGEX-4T-1菌体沉淀; 泳道6. IPTG诱导的pGEX-4T-PtoAED3菌体沉淀。B. 利用GST标签蛋白纯化GST-PtoAED3蛋白的SDS-PAGE电泳图;泳道1 ~ 3分别为洗脱5、3、1次后,谷胱甘肽琼脂糖凝胶偶联物通过SDS-PAGE检测。C. 抗体anti-PtoAED3与纯化后的原核表达蛋白免疫杂交;泳道1. 抗体与IPTG诱导的pGEX-4T-1原核表达蛋白杂交; 泳道2. 抗体与IPTG诱导的pGEX-4T-PtoAED3原核表达蛋白杂交。红色箭头表示原核表达的目的蛋白。A, SDS-PAGE electrophoresis of PtoAED3 prokaryotic expression; lane 1, non-IPTG induced pGEX-4T-1 protein expression in supernatant; lane 2, non-IPTG induced pGEX-4T-PtoAED3 protein expression in supernatant; lane 3, IPTG induced pGEX-4T-1 protein expression in supernatant; lane 4, IPTG induced pGEX-4T-PtoAED3 protein expression in supernatant; lane 5, IPTG induced pGEX-4T-1 protein expression in precipitation; lane 6, IPTG induced pGEX-4T-PtoAED3 protein expression in precipitation. B, SDS-PAGE electrophoresis results using GST-tagged protein purification for GST-PtoAED3 protein; lane 1−3, the glutathione-sepharose eluted 5,3,1 times are detected by SDS-PAGE. The red arrow indicates the interested proteins expressed in E.coli. C, western blotting of anti-PtoAED3 antibody with the purified prokaryotic expression protein; lane 1, western blotting of anti-PtoAED3 antibody with the IPTG-induced pGEX-4T-1 prokaryotic expression protein; lane 2, western blotting of anti-PtoAED3 antibody with the IPTG pGEX-4T-PtoAED3 protein hybridization. The red arrow indicates the protein of interest expressed prokaryotically.
Figure 3. Expression and purification of PtoAED3 in P. tomentosa
图 4 利用GST-pull down在毛白杨中筛选与GST-PtoAED3互作的蛋白
A. 毛白杨植株全蛋白电泳图; B. 还原型谷胱甘肽洗脱与GST-PtoAED3互作的蛋白。泳道1和2为未洗脱的植株全蛋白,泳道3和4分别为利用还原型谷胱甘肽洗脱3和5次收集的蛋白。A, electrophoresis of whole protein of P. tomentosa; B, GST-PtoAED3 interacting protein eluted with reduced glutathione. Lane 1 and 2, uninteracting proteins; lane 3 and 4, proteins with reduced glutathione detected by SDS-PAGE for 3 and 5 times.
Figure 4. Screening of proteins interacting with GST-PtoAED3 in P. tomentosa by GST-pull down
图 5 PtoAED3互作蛋白GO功能富集
Cellular processes 生物学途径包括细胞进程; Metabolism process 代谢过程; Response to stimulus 应激反应; Biological regulation 生物调节; Multicellular organismal process 多细胞器官进程; Regulation of biological process 生物进程调节; Development process 发育过程; Cellular component organization or biogenesis 细胞组分组成或发育; Localization 定位; Reproductive process 繁殖过程; Positive regulation of biological process 生物过程正调节; Multi-organism process 多器官进程; Signaling 信号途径; Negative regulation of biological process 生物过程负调节; Rhythmic process 节律进程; Growth 生长; Immune system process 免疫体统进程; Pigmentation 色素沉积; Biological adhesion 生物固定; Carbon utilization 碳利用; Binding 分子功能包括结合; Catalytic activity 催化活性; Transporter activity 转运活性; Transcription regulator activity 转录调节因子活性; Structural molecular activity 结构分子活性; Antioxidant activity 抗氧化活性; Cell part 细胞组分则包括细胞部分; Cell 细胞; Organelle 细胞器; Membrane 膜; Membrane part 膜组分; Organelle part 细胞器组分; Protein-containing complex 蛋白复合体; Membrane-enclosed lumen 膜封闭腔; Cell junction 细胞连接; Symplast 共质体; Extracellular region 胞外域; Supramolecular complex 超分子复合体; Synapse 突触; Synaptic part and extracellular region part 突触局部及胞外域局部
Figure 5. GO function enrichment of PtoAED3IPs
表 1 GST-pull down筛选的PtoAED3互作蛋白
Table 1 PtoAED3 interacting proteins screened by GST-pull down
项目
Item蛋白编号
Protein No.肽段
Peptide segment蛋白注释
Protein annotation肽段得分
Peptide score覆盖率
Coverage/%高丰度蛋白
High abundance
proteinQ8LPG9 K.EDVEALWLK.K/
R.SQAIIK.LProtein IQ-DOMAIN 14
蛋白IQ结构域20.84/21.91 3.31 Q56XU4 R.MKM*DR.E Zinc finger CCCH domain-containing protein 6
锌指结构域26.92 1.15 C6E2H7 K.KLEWYEK.F GTPase Era
GTP 水解酶27.89 1.99 F4I0N3 R.ELQQLK.E WEB family protein
WEB 转录因子39.89 2.91 O04590 R.VAAEALIR.L Pentatricopeptide repeat-containing protein
三角状五肽23.91 4.88 蛋白质加工降解
Protein processing
and degradationQ9D832 K.NPVNK.K DnaJ homolog subfamily B member 4
分子伴侣21.05 1.58 P43297 K.EGAVAEVK.D Cysteine proteinase RD21A
半胱氨酸蛋白酶20.47 1.72 Q9C5D2 R.MQENISR.F F-box/LRR-repeat protein 4
SCF复合体亚基25.39 1.94 Q9FZK1 K.ILGGKATWATTK.C F-box only protein 6
SCF 复合体亚基21.78 3.22 Q94AH6 R.LLFDK.S Cullin-1 SCF 复合体亚基 37.5 1.22 Q94F30 K.FTLSR.V Ubiquitin-like-specific protease ESD4
泛素特异蛋白酶22.41 0.97 Q9JIG7 R.QNLGKAK.L Coiled-coil domain-containing protein 22
螺旋卷曲结构域蛋白40.1 1.67 O22993 R.FTLSR.S Probable inactive ATP-dependent zinc
metalloprotease FTSHI 1
金属蛋白酶22.41 0.62 胁迫响应相关
Stress response
correlationP43297 K.EGAVAEVK.D Cysteine proteinase RD21A
半胱氨酸蛋白酶20.47 1.72 O80396 M.AGLEELK.K Mitogen-activated protein kinase kinase 3
分裂原活化蛋白激酶27.02 1.33 O04203 R.LPELR.K Nematode resistance protein-like HSPRO2
类线虫抗性蛋白24.23 5.38 Q9LII8 R.IDSELR.H Protein KINESIN LIGHT CHAIN-RELATED 2
驱动蛋白轻链30.55 3.61 O81825 K.LTELR.K Probable disease resistance protein At4g27220
抗病蛋白26.75 0.83 Q9LFP7 R.NVGLK.T Probable serine/threonine-protein kinase PIX7
丝氨酸/苏氨酸蛋白激酶21.76 1.13 Q7XA40 R.SLDELK.R Putative disease resistance protein RGA3
抗病蛋白28.92 7.69 Q9LRR4 R.LLEIR.A Putative disease resistance RPP13-like protein 1
抗病蛋白28.42 1.79 Q9LX93 K.LPRSR.M E3 ubiquitin-protein ligase RING1
E3 泛素连接酶22.21 17.86 Q7VG78 K.AGPLSARKPASK.Q Probable GMP synthase [glutamine-hydrolyzing]
GMP合成酶22.29 23.53 Q9C5D2 R.MQENISR.F F-box/LRR-repeat protein 4
SCF 复合体亚基25.39 1.94 Q0WQF4 K.ITELR.E Vacuolar protein sorting-associated protein 53 A
液泡蛋白分类相关蛋白26.75 1.48 O22769 R.LLEIR.Q NADH dehydrogenase [ubiquinone] flavoprotein 2
NADH 脱氢酶[泛醌]黄素蛋白28.42 1.97 Q9UI10 K.IIIADK.V Translation initiation factor eIF-2B subunit delta
翻译起始因子亚基25.12 0.94 O65202 R.ILELR.L Peroxisomal acyl-coenzyme A oxidase 1
过氧化物酶体酰基辅酶A氧化酶28.42 0.75 F4I9E1 R.LKM*IPR.T Protein NUCLEAR FUSION DEFECTIVE 4
核融合缺陷蛋白25.57 1.06 P47140 R.EQSNVIAR.Q Altered inheritance rate of mitochondria protein 25
线粒体蛋白21.26 2.31 O43099 R.LPELR.K Peroxiredoxin Asp f3
过氧化物酶24.23 4.13 Q9FYA2 K.ENYHLVPR.T Probable WRKY transcription factor 75
WRKY转录因子32.11 6.35 Q84X53 R.SLDELK.E Transcription termination factor MTEF1, chloroplastic
转录终止因子28.92 2.48 发育过程相关
Developmental
processQ9UI10 K.IIIADK.V Translation initiation factor eIF-2B subunit delta
翻译起始因子亚基25.12 0.94 Q84JG2 K.ILRFEELDLQM*EK.E SWI/SNF complex subunit SWI3B
SWI/SNF复合体亚基23.07 2.60 Q12955 K.AGMTPLDLATNEEIR.L Ankyrin-3 锚蛋白 24.76 8.72 Q8GWQ6 K.AYALDK.K UPF0235 protein At5g63440 UPF蛋白 21.38 6.25 Q84X53 R.SLDELK.E Transcription termination factor MTEF1, chloroplastic
转录终止因子28.92 2.48 F4JRR5 R.VDTFR.I TITAN-like protein
TITAN蛋白21.33 3.03 Q94AH6 R.LLFDK.S Cullin-1
SCF 复合体亚基37.5 1.22 O22993 R.FTLSR.S Probable inactive ATP-dependent zinc metalloprotease FTSHI 1
金属蛋白酶22.41 0.62 Q9LII8 R.IDSELR.H Protein KINESIN LIGHT CHAIN-RELATED 2
驱动蛋白轻链30.55 3.61 Q0D4T4 K.VELKR.I MADS-box transcription factor 18
MADS转录因子23.86 2.53 Q9ZVP8 K.VLEQR.A NAC domain-containing protein 35
NAC转录因子25.43 1.98 Q94F30 K.FTLSR.V Ubiquitin-like-specific protease ESD4
泛素特异蛋白酶22.41 0.97 Q9FYA2 K.ENYHLVPR.T Probable WRKY transcription factor 75
WRKY转录因子32.11 6.35 Q9FII5 R.LGGMIPAK.T Leucine-rich repeat receptor-like protein kinase TDR
受体蛋白激酶23.78 5.88 Q9LHP4 R.IPELR.T Receptor-like protein kinase 2
受体蛋白激酶24.23 3.57 Q9SN39 R.ILFDK.Y Pentatricopeptide repeat-containing protein DOT4
三角状五肽蛋白37.5 2.53 F4I9E1 R.LKM*IPR.T Protein NUCLEAR FUSION DEFECTIVE 4
核融合缺陷蛋白25.57 1.06 Q8LEZ4 R.AGVERK.A Protein NUCLEAR FUSION DEFECTIVE 5
核融合缺陷蛋白25.82 0.84 Q9FZK1 K.ILGGKATWATTK.C F-box only protein 6
SCF 复合体亚基21.78 3.22 Q9M811 K.EAPQVPASPR.E Rop guanine nucleotide exchange factor 11
鸟苷酸交换因子21.47 7.41 Q2QDF6 .LSEVR.G Steroid 5-alpha-reductase DET2
类固醇还原酶22.75 6.33 -
[1] 葛伟娜, 李超, 张家琦. 植物天冬氨酸蛋白酶的研究进展[J]. 生物技术通报, 2016, 282(1):14−20. Ge W N, Li C, Zhang J Q. Research advances on plant aspartic proteinase[J]. Biotechnology Bulletin, 2016, 282(1): 14−20.
[2] 王亚锐, 吴燕. 植物天冬氨酸蛋白酶的功能研究进展[J]. 生命科学, 2016, 204(3):106−112. Wang Y R, Wu Y. The research progress on the functions of plant aspartic proteases[J]. Chinese Bulletin of Life Sciences, 2016, 204(3): 106−112.
[3] Suguna K, Padlan E A, Smith C W, et al. Binding of a reduced peptide inhibitor to the aspartic proteinase from Rhizopus chinensis: implications for a mechanism of action[J]. Proceedings of the National Academy of Sciences of the United States of America, 1987, 84(20): 7009−7013. doi: 10.1073/pnas.84.20.7009
[4] Zhao G H, Zhou A H, Lu G, et al. Identification and characterization of Toxoplasma gondii aspartic protease 1 as a novel vaccine candidate against toxoplasmosis[J/OL]. Parasites & Vectors, 2013, 6: 175 (2013−06−14)[2019−05−26]. https://doi.org/10.1186/1756-3305-6-175.
[5] Takahashi K, Niwa H, Yokota N, et al. Widespread tissue expression of nepenthesin-likeaspartic protease genes in Arabidopsis thaliana[J]. Plant Physiology and Biochemistry, 2008, 46(7): 724−729. doi: 10.1016/j.plaphy.2008.04.007
[6] Chen J, Ouyang Y, Wang L, et al. Aspartic proteases gene family in rice: gene structure and expression, predicted protein features and phylogenetic relation[J]. Gene, 2009, 442(1−2): 108−118. doi: 10.1016/j.gene.2009.04.021
[7] Guo R, Xu X, Carole B, et al. Genome-wide identification, evolutionary and expression analysis of the aspartic protease gene superfamily in grape[J/OL]. BMC Genomics, 2013, 14: 554 (2013−08−15)[2019−06−27]. https://doi.org/10.1186/1471-2164-14-554.
[8] 高杉, 蓝兴国. 植物天冬氨酸蛋白酶的结构与功能[J]. 生物技术通讯, 2018, 29(6):150−154. Gao S, Lan X G. Structure and function of aspartic proteinases in plants[J]. Letters in Biotechnology, 2018, 29(6): 150−154.
[9] Faro C, Gal S. Aspartic proteinase content of the Arabidopsis genome[J]. Current Protein Peptide Science, 2005, 6(6): 493−500. doi: 10.2174/138920305774933268
[10] Simoes I, Faro C. Structure and function of plant aspartic proteinases[J]. European Journal of Biochemistry, 2004, 271(11): 2067−2075. doi: 10.1111/j.1432-1033.2004.04136.x
[11] Runeberg-Roos P, Saarma M. Phytepsin, a barley vacuolar aspartic proteinase, is highly expressed during autolysis of developing tracheary elements and sieve cells[J]. Plant Journal, 1998, 15(1): 139−145. doi: 10.1046/j.1365-313X.1998.00187.x
[12] Tamura T, Terauchi K, Kiyosaki T, et al. Differential expression of wheat aspartic proteinases, WAP1 and WAP2, in germinating and maturing seeds[J]. Plant Physiology, 2007, 164(4): 470−477. doi: 10.1016/j.jplph.2006.02.009
[13] Kato Y, Yamamoto Y, Murakami S, et al. Post-translational regulation of CND41 protease activity in senescent tobacco leaves[J]. Planta, 2005, 222(4): 643−651. doi: 10.1007/s00425-005-0011-4
[14] Diaz C, Lemaitre T, Christ A, et al. Nitrogen recycling and remobilization are differentially controlled by leaf senescence and development stage in Arabidopsis under low nitrogen nutrition[J]. Plant Physiology, 2008, 147(3): 1437−1449. doi: 10.1104/pp.108.119040
[15] Ge X, Dietrich C, Matsuno M, et al. An Arabidopsis aspartic protease functions as an anti-cell-death component in reproduction and embryogenesis[J]. Embo Reports, 2005, 6(3): 282−288. doi: 10.1038/sj.embor.7400357
[16] Bright J, Desikan R, Hancock J T, et al. ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis[J]. Plant Journal, 2006, 45(1): 113−122. doi: 10.1111/j.1365-313X.2005.02615.x
[17] Xia Y, Suzuki H, Borevitz J, et al. An extracellular aspartic protease functions in Arabidopsis disease resistance signaling[J]. Embo Journal, 2004, 23(4): 980−988. doi: 10.1038/sj.emboj.7600086
[18] Cao S, Guo M, Wang C, et al. Genome-wide characterization of aspartic protease (AP) gene family in Populus trichocarpa and identification of the potential PtAPs involved in wood formation[J]. BMC Plant Biology, 2019, 19: 1−17.
[19] Bhalerao R, Keskitalo J, Sterky F, et al. Gene expression in autumn leaves[J]. Plant Physiology, 2003, 131(2): 430−442. doi: 10.1104/pp.012732
[20] Rauch J N, Gestwicki J E. Binding of human nucleotide exchange factors to heat shock protein 70 (Hsp70) generates functionally distinct complexes in vitro[J]. Journal of Biological Chemistry, 2014, 289(3): 1402−1414. doi: 10.1074/jbc.M113.521997
[21] Starokadomskyy P, Gluck N, Li H, et al. CCDC22 deficiency in humans blunts activation of proinflammatory NF-κB signaling[J]. Journal of Clinical Investigation, 2013, 123(5): 2244−2256. doi: 10.1172/JCI66466
[22] Murtas G, Reeves P H, Fu Y F, et al. A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of SMALL UBIQUITIN-RELATED MODIFIER conjugates[J]. Plant Cell, 2003, 15(10): 2308−2319. doi: 10.1105/tpc.015487
[23] Shindo T, Misas-Villamil J C, Hörger A C, et al. A role in immunity for Arabidopsis cysteine protease RD21, the ortholog of the tomato immune protease C14[J/OL]. PLoS ONE, 2012, 7(1): e29317 (2012−01−06) [2020−10−10]. https://doi.org/10.1371/journal.pone.0029317.
[24] Lampl N, Alkan N, Davydov O, et al. Set-point control of RD21 protease activity by AtSerpin1 controls cell death in Arabidopsis[J]. Plant Journal, 2013, 74(3): 498−510. doi: 10.1111/tpj.12141
[25] Hayashi Y, Yamada K, Shimada T, et al. A proteinase-storing body that prepares for cell death or stresses in the epidermal cells of Arabidopsis[J]. Plant Cell Physiology, 2001, 42(9): 894−899. doi: 10.1093/pcp/pce144
[26] Dóczi R, Brader G, Pettkó-Szandtner A, et al. The Arabidopsis mitogen-activated protein kinase kinase MKK3 is upstream of group C mitogen-activated protein kinases and participates in pathogen signaling[J]. Plant Cell, 2007, 19(10): 3266−3279. doi: 10.1105/tpc.106.050039
[27] Murray S L, Ingle R A, Petersen L N, et al. Basal resistance against Pseudomonas syringae in Arabidopsis involves WRKY53 and a protein with homology to a nematode resistance protein[J]. Molecular Plant Microbe Interact, 2007, 20(11): 1431−1438. doi: 10.1094/MPMI-20-11-1431
[28] Lin S S, Martin R, Mongrand S, et al. RING1 E3 ligase localizes to plasma membrane lipid rafts to trigger FB1-induced programmed cell death in Arabidopsis[J]. Plant Journal, 2008, 56(4): 550−561. doi: 10.1111/j.1365-313X.2008.03625.x
[29] Wang L C, Tsai M C, Chang K Y, et al. Involvement of the Arabidopsis HIT1/AtVPS53 tethering protein homologue in the acclimation of the plasma membrane to heat stress[J]. Journal of Experimental Botnay, 2011, 62(10): 3609−3620. doi: 10.1093/jxb/err060
[30] Lee C F, Pu H Y, Wang L C, et al. Mutation in a homolog of yeast Vps53p accounts for the heat and osmotic hypersensitive phenotypes in Arabidopsis hit1-1
mutant[J]. Planta, 2006, 224(2): 330−338. doi: 10.1007/s00425-005-0216-6 [31] Hillmann F, Bagramyan K, Straßburger M, et al. The crystal structure of peroxiredoxin Asp f3 provides mechanistic insight into oxidative stress resistance and virulence of Aspergillus fumigatus[J/OL]. Scientific Reports, 2016, 6(1): 33396 (2016−09−14) [2019−08−03]. https://doi.org/10.1186/1471-2164-14-554.
[32] Meskauskiene R, Würsch M, Laloi C, et al. A mutation in the Arabidopsis mTERF-related plastid protein SOLDAT10 activates retrograde signaling and suppresses 1O2-induced cell death[J]. Plant Journal, 2009, 60(3): 399−410. doi: 10.1111/j.1365-313X.2009.03965.x
[33] Sarnowski T J, Ríos G, Jásik J, et al. SWI3 subunits of putative SWI/SNF chromatin-remodeling complexes play distinct roles during Arabidopsis development[J]. Plant Cell, 2005, 17(9): 2454−2472. doi: 10.1105/tpc.105.031203
[34] Lu X, Li Y, Su Y, et al. An Arabidopsis gene encoding a C2H2-domain protein with alternatively spliced transcripts is essential for endosperm development[J]. Journal of Experimental Botany, 2012, 63(16): 5935−5944. doi: 10.1093/jxb/ers243
[35] Shen W H, Parmentier Y, Hellmann H, et al. Null mutation of AtCUL1 causes arrest in early embryogenesis in Arabidopsis[J]. Molecular Biology of the Cell, 2002, 13(6): 1916−1928. doi: 10.1091/mbc.e02-02-0077
[36] Hobbie L, McGovern M, Hurwitz L R, et al. The axr6
mutants of Arabidopsis thaliana define a gene involved in auxin response and early development[J]. Development, 2000, 127(1): 23−32. [37] Song J B, Huang S Q, Dalmay T, et al. Regulation of leaf morphology by microRNA394 and its target LEAF CURLING RESPONSIVENESS[J]. Plant & Cell Physiology, 2012, 53(7): 1283−1294.
[38] Kadirjan-Kalbach D K, Yoder D W, Ruckle M E, et al. FtsHi1/ARC1 is an essential gene in Arabidopsis that links chloroplast biogenesis and division[J]. Plant Journal, 2012, 72(5): 856−867. doi: 10.1111/tpj.12001
[39] Fornara F, Parenicová L, Falasca G, et al. Functional characterization of OsMADS18
, a member of the AP1/SQUA subfamily of MADS box genes[J]. Plant Physiology, 2004, 135(4): 2207−2219. doi: 10.1104/pp.104.045039 [40] Yoo S Y, Kim Y, Kim S Y, et al. Control of flowering time and cold response by a NAC-domain protein in Arabidopsis[J/OL]. PLoS ONE, 2007, 2(7): e642(2007−07−25)[2020−10−12]. https://doi.org/10.1371/journal.pone.0000642.
[41] Devaiah B N, Karthikeyan A S, Raghothama K G. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis[J]. Plant Physiology, 2007, 143(4): 1789−1801. doi: 10.1104/pp.106.093971
[42] Shin D H, Cho M H, Kim T L, et al. A small GTPase activator protein interacts with cytoplasmic phytochromes in regulating root development[J]. Journal of Biological Chemistry, 2010, 285(42): 32151−32159. doi: 10.1074/jbc.M110.133710
-
期刊类型引用(4)
1. 杨桦,李祥乾,王帆,方睿,杨伟. 长足大竹象信息素结合蛋白CbuqPBP2互作蛋白的筛选与验证. 西北农林科技大学学报(自然科学版). 2024(01): 87-97 .  百度学术
百度学术
2. 万超,张月,胡莉,伍炳华,袁媛. 茉莉花JsMYB305转录因子的原核表达及蛋白纯化. 福建农业学报. 2022(02): 164-169 .  百度学术
百度学术
3. 武建颖,张燕,孙贺贺,赵玉兰,董金皋,申珅,郝志敏. 玉米大斑病菌蛋白激酶A催化亚基StPKA-C1/C2的表达与互作蛋白筛选. 农业生物技术学报. 2022(10): 1976-1986 .  百度学术
百度学术
4. 胡莉,万超,张蕖,陈清西,伍炳华,袁媛. 茉莉花JsMYB305转录因子互作蛋白的筛选及验证. 福建农业学报. 2022(09): 1135-1144 .  百度学术
百度学术
其他类型引用(9)




 下载:
下载: