Cloning and expression pattern analysis of FmPIF gene family in Fraxinus mandshurica
-
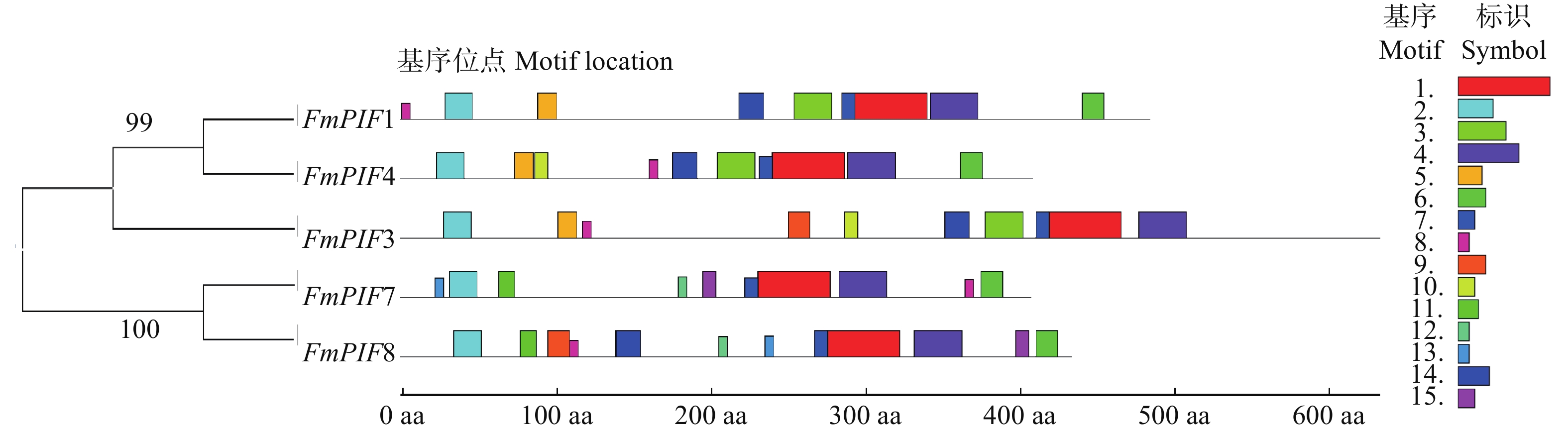
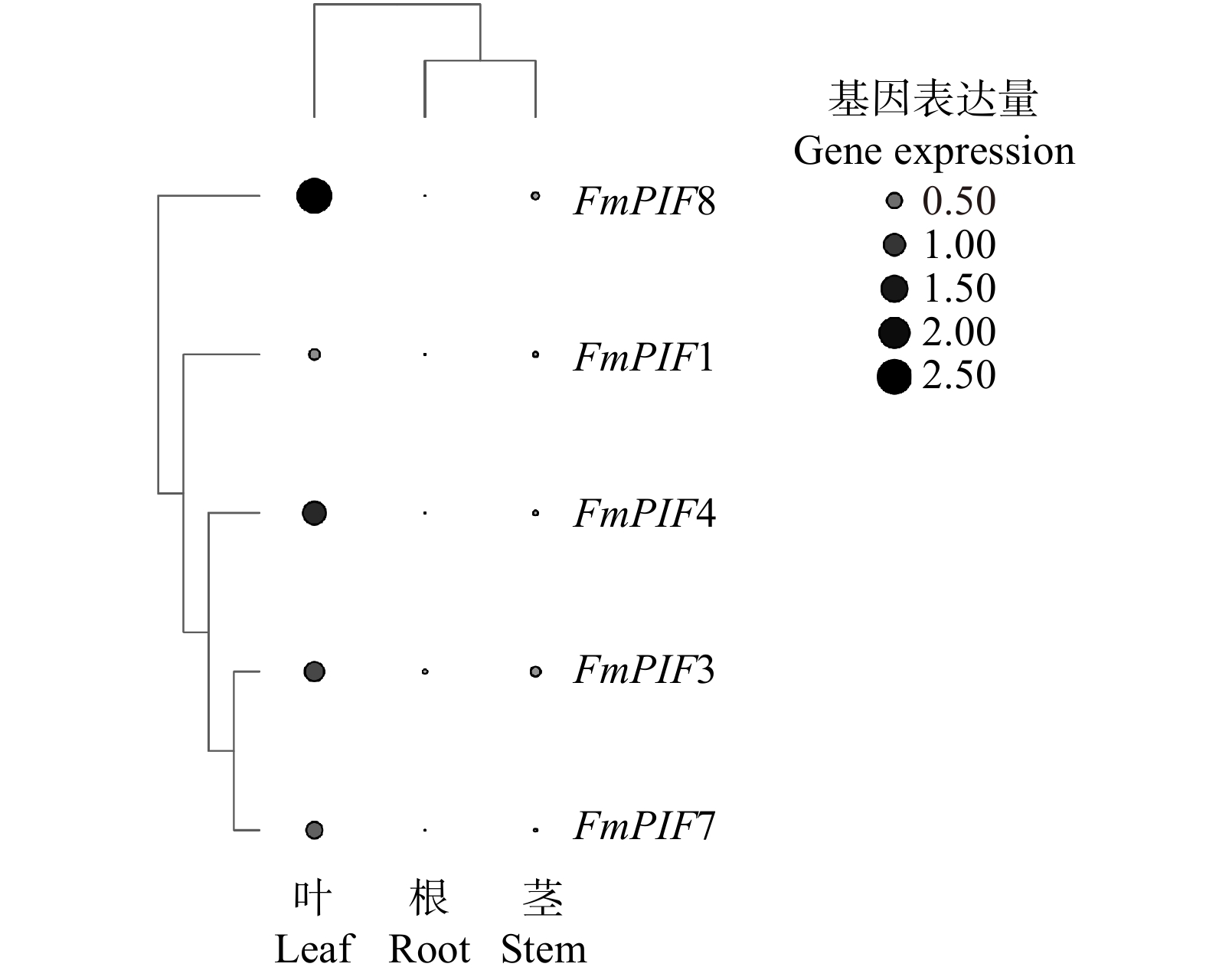
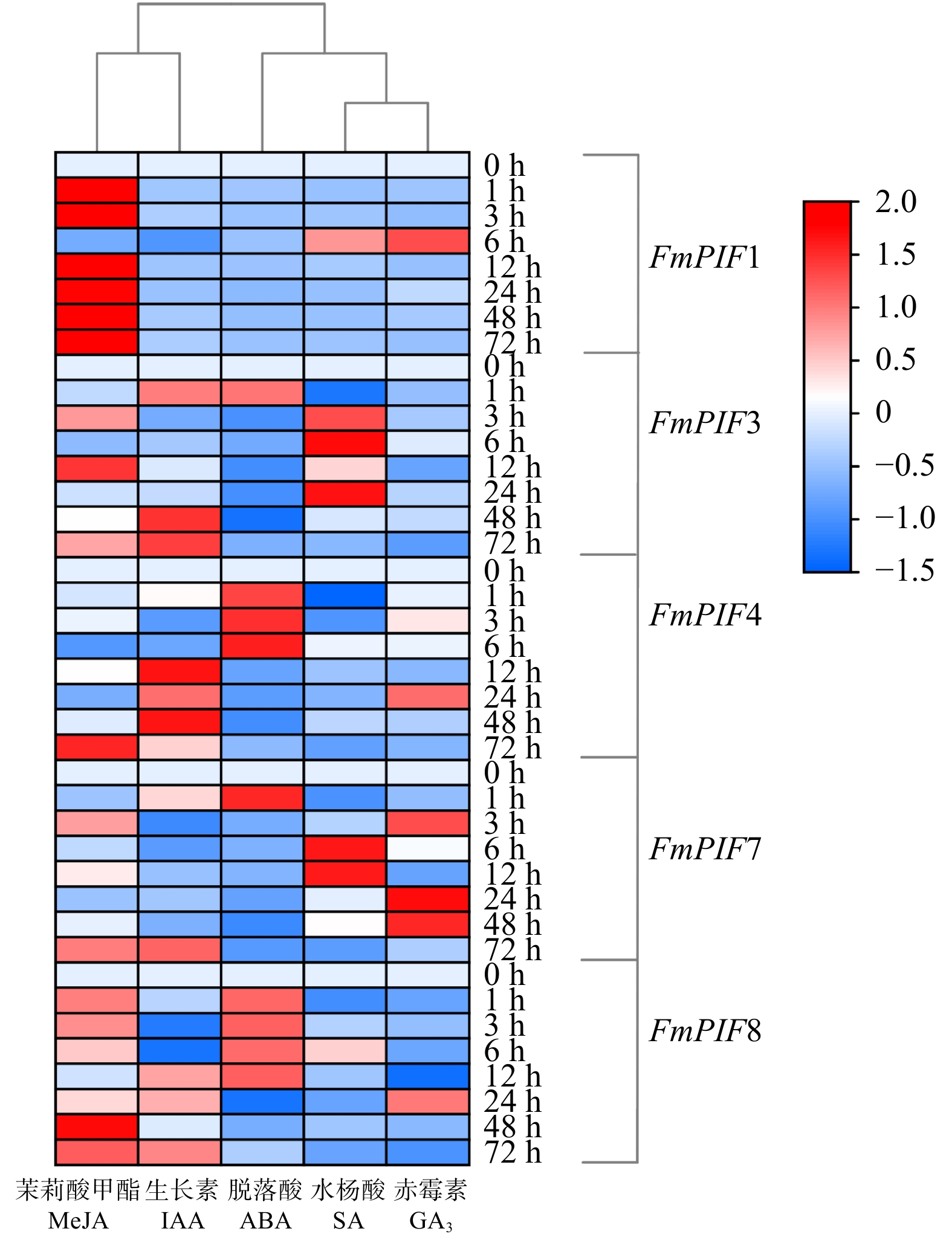
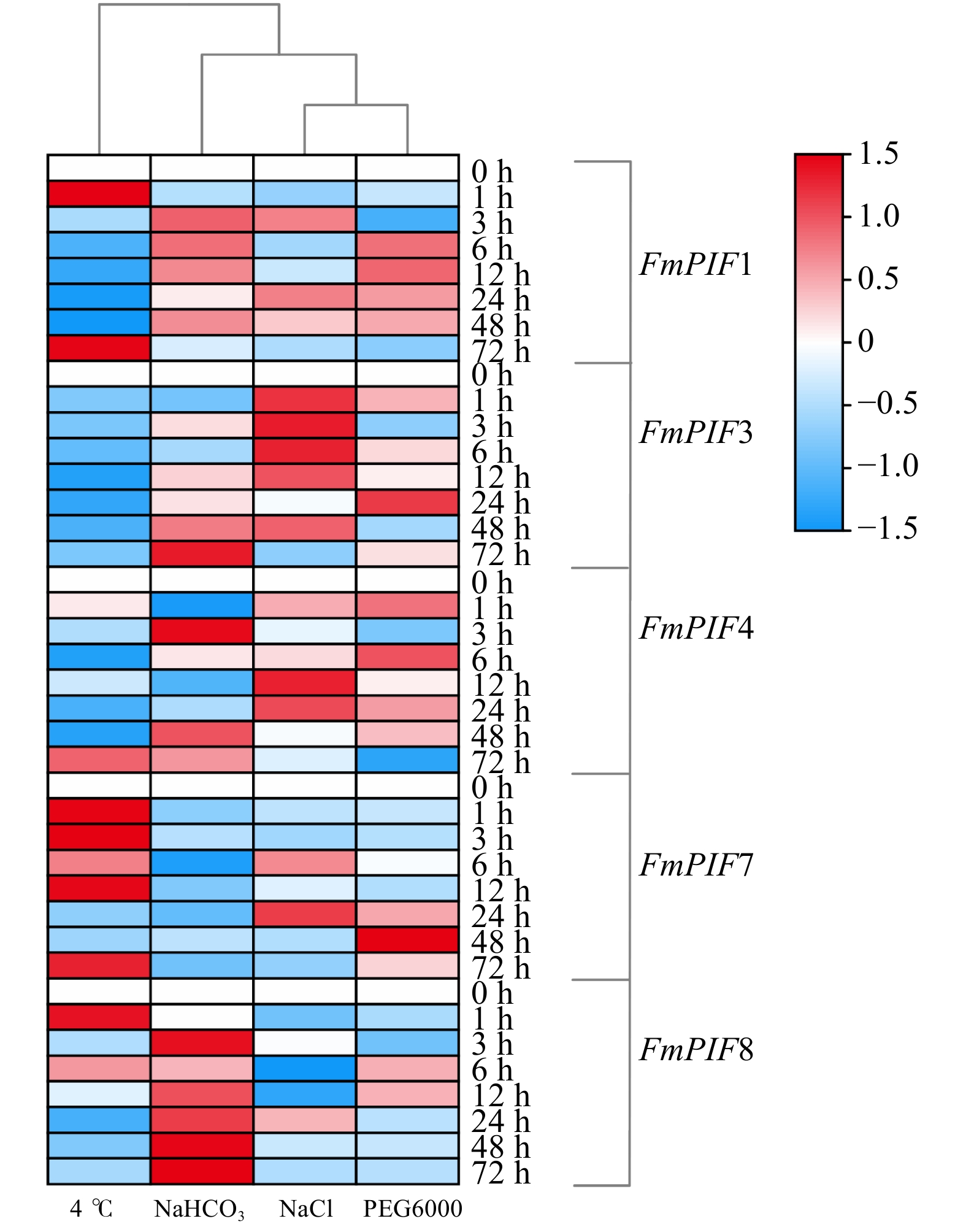
摘要:目的 探究水曲柳光敏色素互作因子(FmPIFs)在激素调控与非生物胁迫应答过程中的重要作用,为揭示水曲柳抗逆分子机制和制定林木遗传育种策略提供理论依据。方法 在水曲柳中克隆FmPIFs基因,并对其基因结构、蛋白质理化性质、保守基序、系统进化关系等进行生物信息学分析。采用qRT-PCR方法分析水曲柳中FmPIFs基因在不同组织及不同激素与胁迫条件下的表达模式。结果 获得5个水曲柳FmPIFs基因家族成员,分别命名为FmPIF1、FmPIF3、FmPIF4、FmPIF7和FmPIF8,其对应的蛋白均为亲水性不稳定蛋白,全部定位在细胞核内。多序列比对结果表明FmPIFs均存在APB保守结构域,成员FmPIF1与FmPIF3存在特有的APA结构域。组织特异性分析显示FmPIFs均在叶中表达量最高,其中成员FmPIF8表达量最高,为对照的3.96倍;但在茎中少量表达,茎中表达量最高的是FmPIF3,仅是对照的0.21倍;而在根中表达量均极低。胁迫响应分析表明,FmPIFs正调控水曲柳植株盐、碱和干旱胁迫抗性,而对植株抗寒性起负调控作用,其中成员FmPIF3对寒冷及盐胁迫明显响应,FmPIF8在碱胁迫下表达量明显上调,FmPIF1在干旱胁迫下表达量明显上调。在激素响应结果中,FmPIFs对脱落酸(ABA)、水杨酸(SA)和赤霉素(GA3)的响应较为一致,而对生长素(IAA)和茉莉酸甲酯(MeJA)的响应存在差异。FmPIF1在施加MeJA后剧烈应答且表达量显著上调,FmPIF7在SA处理后表达量明显上调,FmPIF3和FmPIF4在GA3处理后表达量明显上调。结论 FmPIFs各成员在基因及蛋白结构上表现出较高的一致性。RT-qPCR结果表明,FmPIFs在水曲柳叶片中表达量最高。在盐、碱、干旱和寒冷胁迫下,FmPIFs被诱导表达,且大部分表达模式相似。FmPIFs也在水曲柳响应IAA、ABA、MeJA、SA、GA3激素调控过程中发挥重要作用。Abstract:Objective This paper aims to explore the important role of Fraxinus mandshurica phytochrome interaction factors (PIFs) in the process of hormone regulation and abiotic stress response, and provide theoretical basis for revealing the molecular mechanism of Fraxinus mandshurica resistance and formulating forest genetic breeding strategies.Method The FmPIFs gene was cloned from Fraxinus mandshurica, and its gene structure, protein physicochemical properties, conserved motifs, and phylogenetic relationships were analyzed by bioinformatics. The qRT-PCR method was used to analyze the expression patterns of FmPIFs genes in Fraxinus mandshurica in different tissues and under different hormones and stress conditions.Result Five members of FmPIFs gene family of Fraxinus mandshurica were obtained and named as FmPIF1, FmPIF3, FmPIF4, FmPIF7 and FmPIF8. The corresponding proteins were all hydrophilic and unstable proteins, all of which were located in the nucleus. The results of multiple sequence alignment showed that FmPIFs all had APB conserved domains, and members FmPIF1 and FmPIF3 had unique APA domains. Tissue-specific analysis showed that FmPIFs were all expressed in leaves at the highest level, and member FmPIF8 expressed at the highest level, which was 3.96 times of control. However, it was expressed in a small amount in the stem, and the highest expression in stem was FmPIF3, which was only 0.21 time of control. The expression in root was extremely low. Stress response analysis showed that FmPIFs positively regulated the resistance of Fraxinus mandshurica plants to salt, alkali and drought stress, while negatively regulated plant cold resistance. The member FmPIF3 responded significantly to cold and salt stress, and the expression of FmPIF8 was significantly up-regulated under alkali stress, the expression of FmPIF1 was significantly up-regulated under drought stress. In the hormone response results, FmPIFs hadrelatively consistent responses to abscisic acid (ABA), salicylic acid (SA) and gibberellin (GA3), while responses to auxin (IAA) and methyl jasmonate (MeJA) existed difference. FmPIF1 responded violently after MeJA application and its expression was significantly up-regulated, FmPIF7 was significantly up-regulated after SA treatment, and FmPIF3 and FmPIF4 were significantly up-regulated after GA3 treatment.Conclusion FmPIFs show high consistency in gene and protein structure. RT-qPCR results show that FmPIFs express the highest amount in the leaves of Fraxinus mandshurica. FmPIFs are induced to express by salt, alkali, drought and cold stress, and most of the expression patterns are similar. FmPIFs also play an important role in the regulation of Fraxinus mandshurica in response to IAA, ABA, MeJA, SA and GA3 hormones.
-
《北京林业大学学报》(原名《北京林学院学报》)创刊于1979年,由教育部主管、北京林业大学主办,国内外公开发行。历任主编分别为我国6位著名林学家汪振儒、沈国舫、关毓秀、王九龄、贺庆棠、尹伟伦。
《北京林业大学学报》是中文核心期刊、中国科技核心期刊、中国科学引文数据库统计源期刊、中国科技论文统计源期刊。荣获第二届国家期刊奖提名奖、第三届国家期刊奖百种重点期刊、中国精品科技期刊、中国高校精品科技期刊、中国国际影响力优秀学术期刊、“中国科技论文在线优秀期刊”一等奖等。
连续收录《北京林业大学学报》的著名检索期刊和数据库有:美国《化学文摘》(CA)、俄罗斯《文摘杂志》(AJ)、英国国际农业与生物学数据库(CABI)、英国《动物学记录》(ZR)、中国科学引文数据库(CSCD)、中国科技论文统计与引文分析数据库(CSTPCD)、《中国学术期刊文摘》《中国生物学文摘》、中国林业科技文献数据库等。
《北京林业大学学报》是中国最有代表性的林业科学期刊之一,主要刊登代表中国林业科学研究前沿创新水平的稿件。期刊定位为“立足中国,面向世界”的全国性林业科学期刊。面向国内外作者广泛征稿,对校内外稿件的质量要求一视同仁。
为保持学科特色,《北京林业大学学报》重点报道以林木遗传育种学、森林培育学、森林经理学、森林生态学、树木生理学、森林土壤学、森林植物学、森林保护学、自然保护区学、园林植物与观赏园艺、风景园林、水土保持与荒漠化防治、森林工程、木材科学与技术、林产化学加工工程、其他学科在林学上的应用等方面的论文。
《北京林业大学学报》现拥有以北京林业大学、中国林业科学研究院、中国科学院、国内其他综合性大学、农林院校、工科院校以及国外有关科研机构和大学等单位的研究人员为主的作者队伍。近年来随着期刊学术水平和影响因子的不断提高,投稿量显著增加,其中校外作者的投稿量占总收稿量的2/3左右。在此,我们对所有给《北京林业大学学报》赐稿的作者表示衷心的感谢!
《北京林业大学学报》自2015年起由原来的双月刊改为单月刊,大16开本,每月月底出版。每期定价50元。各地邮局发行,邮发代号:82−304。国内统一刊号:CN 11−1932/S。如当地邮局订阅不便或错过征订时间,也可直接汇款向本刊编辑部订阅。
地址:北京市海淀区清华东路35号《北京林业大学学报》编辑部
邮编:100083 发行电话:010−62338397 联系人:刘大林
发行电子信箱:liudalin@bjfu.edu.cn
网址:http://j.bjfu.edu.cn,http://journal.bjfu.edu.cn
-
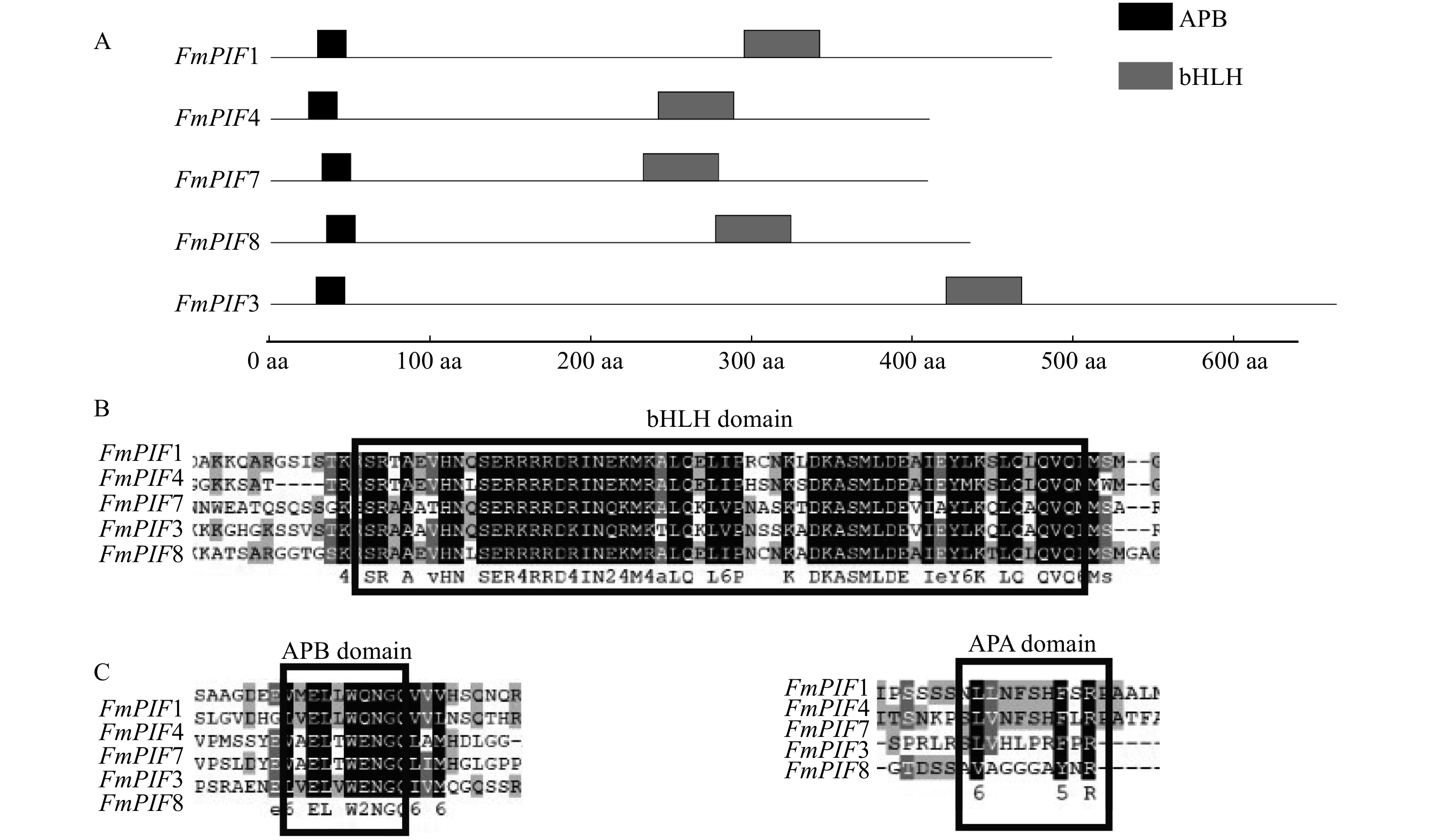
图 2 水曲柳FmPIFs基因保守结构域
A. 水曲柳FmPIFs基因保守结构域位置;B. 水曲柳FmPIFs基因bHLH保守结构域;C. FmPIFs基因的APA和APB保守结构域。A, conservative domain location of FmPIFs gene in F. mandshurica; B, bHLH domain of FmPIFs gene in F. mandshurica; C, APA and APB domain of FmPIFs gene in F. mandshurica.
Figure 2. Conserved domain of FmPIFs gene in F. mandshurica
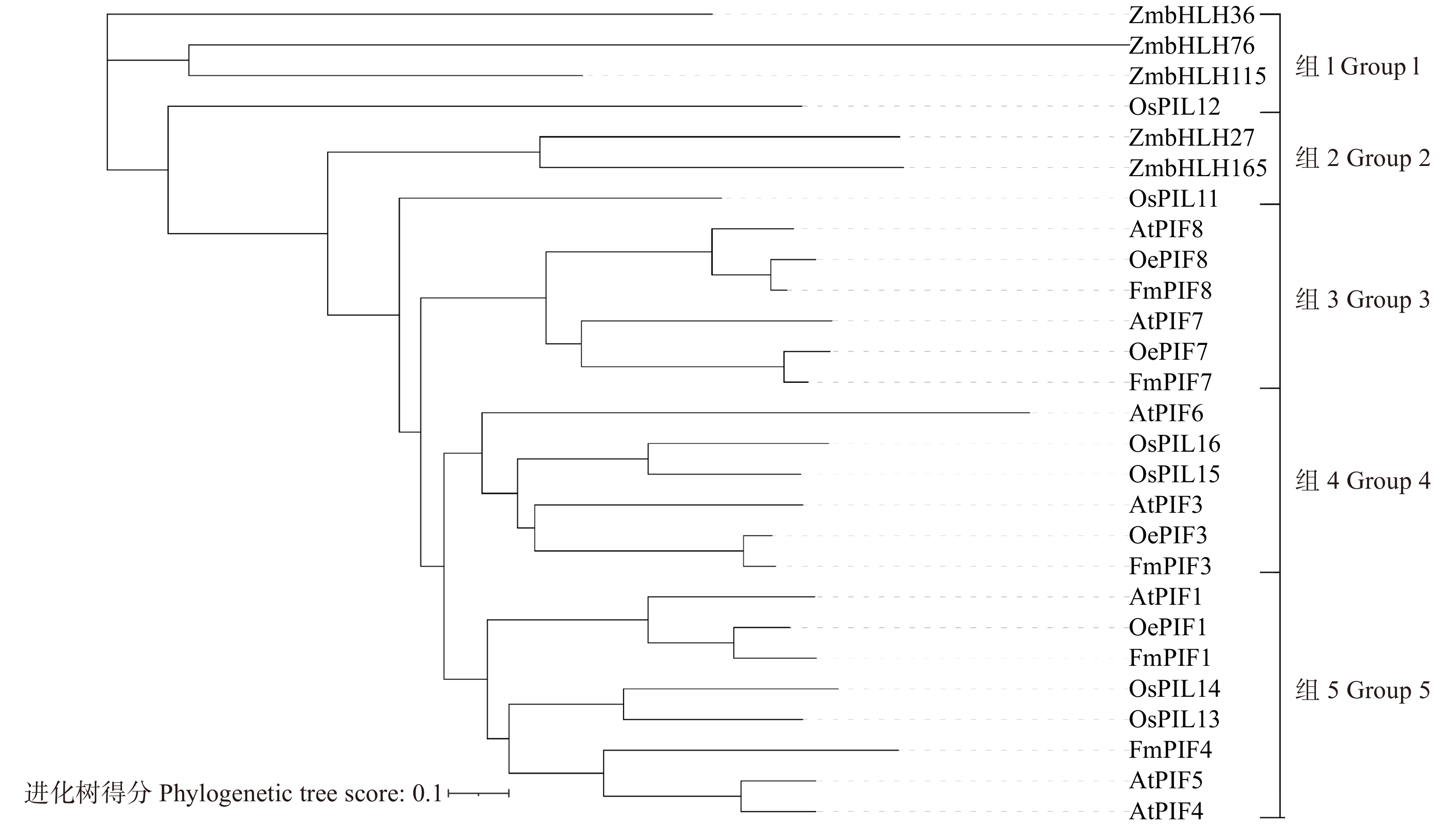
图 3 水曲柳系统进化树分析
AtPIF1. AT2G20180;AtPIF3. AT1G09530;AtPIF4. AT2G43010;AtPIF5. AT3G59060;AtPIF6. AT3G62090;AtPIF7. AT5G61270; AtPIF8. AT4G00050;OsPIL11. Os12g0610200;OsPIL12. Os03g0639300;OsPIL13. Os03g0782500;OsPIL14. Os07g0143200; OsPIL15. Os01g0286100;OsPIL16. Os05g0139100;OePIF1. XP_022874602.1;OePIF3. XP_022861889.1;OePIF7. XP_022885445.1;OePIF8. XP_022897201.1;ZmbHLH27. AIB04350.1;ZmbHLH36. AIB04280.1;ZmbHLH76. AIB04359.1;ZmbHLH115. AIB04374.1;ZmbHLH165. AIB04376.1。
Figure 3. Phylogenetic tree analysis of F. mandshurica
表 1 克隆FmPIFs基因编码区对应引物
Table 1 Corresponding primers of coding region for cloning FmPIFs gene
引物名称 Primer name 引物序列(5′—3′) Primer sequence(5′−3′) PIF1-F ACTTAATCAGTGAGAGGCACAGAA PIF1-R TTCCCCAGTTGGAATCGGAT PIF3-F TAGGAGTTGTTTCATCGGTGGA PIF3-R ATCAGTTGCATTGGTAGGGGTA PIF4-F TGCTTTCATTTCCAAGGTTC PIF4-R GAGCATTAGAGTCGTCCTACGT PIF7-F GAGAGGAGAAAGAGTCTCTGAGACA PIF7-R GAGAAGCAAAATCAATAAAAATAACG PIF8-F TTGAAGTCTACCTATGTACACTGTGTTT PIF8-R ATTTTGTTGTTGGAACTGTTGATAC 表 2 生物信息学分析软件
Table 2 Software for bioinformatics analysis
生物信息分析
Bioinformatics analysis软件及网址
Software and website核酸序列分析 Nucleic acid sequence analysis DNAMAN 氨基酸理化性质分析
Analysis of physical and chemical properties of amino acidExPASy ProtParam tool[28] https://web.expasy.org/protparam/ 蛋白跨膜结构预测 Protein transmembrane structure prediction TMHMM SERVER 2.0[29] http://www.cbs.dtu.dk/services/TMHMM/ 蛋白亲水/疏水分析 Protein hydrophilic/hydrophobic analysis ProtScale[28] https://web.expasy.org/protscale/ 蛋白结构域分析 Protein domain analysis NCBI Conserved Domain Search Service(CD Search)
https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi TBtools蛋白信号肽预测 Protein signal peptide prediction SignalP 5.0 server[30] http://www.cbs.dtu.dk/services/SignalP/ 亚细胞定位预测 Subcellular location prediction wolf psort[31] https://wolfpsort.hgc.jp/ 蛋白二级结构预测 Protein secondary structure prediction GOR IV https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_gor4.html 基因结构绘制 Gene structure mapping GSDS2.0[32] http://gsds.gao-lab.org/index.php 保守基序分析 Conserved motif analysis MEME Suite[33] http://meme-suite.org/tools/meme 多序列比对 Multiple sequence alignment Clustal X2.0[34] 系统进化树 Phylogenetic tree MEGA5.1[31] 进化树美化 Evolutionary tree beautification ITOL[35] https://itol.embl.de/ 表 3 FmPIFs定量引物
Table 3 Quantitative primers of FmPIFs
引物名称 Primer name 引物序列(5′—3′) Primer sequence(5′−3′) QRT-PIF1-1F TCTTCTCTATTCTACTCCACCTGCTCCA QRT-PIF1-1R TGTGCCGTTGTCGCCTTGCT QRT-PIF3-1F CTCACATGCCAAGAGTTCGAGATGC QRT-PIF3-1R AGACATTCCATTAGCCGTAACACCAGATA QRT-PIF4-1F ACAACATCTTCAGGTGGATCTGGTAGTAG QRT-PIF4-1R CCTCCTTCTCCTCTCCGACAAGTTATG QRT-PIF7-1F TCTTGGTGGCATGGTTCCGTCTC QRT-PIF7-1R CAACTGCGTGTCAACTGATGAACTCTT QRT-PIF8-1F GAATGCCTCCAGTTCTATCTCCTCCTC QRT-PIF8-1R CTGTTGATACAAAGTTGCCATCCTGCTATA α-TubuLin-F GCACTGGCCTCCAAGGAT α-TubuLin-R TGGGTCGCTCAATGTCAAGG 表 4 水曲柳中FmPIFs成员信息
Table 4 Information of FmPIFs members in Fraxinus mandshurica
序列名称
Sequence
nameGenBank登录号
GenBank accession
No.编码区全长
Full length of
coding area/bp氨基酸长度
Amino acid
length/aa等电点
Isoelectric
point蛋白分子质量
Molecular mass
of protein/Da不稳定系数
Instability
coefficient总平均疏水性
Total average
hydrophobicityPIF1 MW268689 1 557 518 6.41 56 644.65 61.95 −0.743 PIF3 MW268690 2 124 707 5.84 75 761.54 56.61 −0.564 PIF4 MW268691 1 314 437 6.66 48 975.67 55.91 −0.864 PIF7 MW268692 1 311 436 8.03 48 102.08 67.10 −0.753 PIF8 MW268693 1 395 464 8.54 49 994.70 57.51 −0.539 表 5 水曲柳FmPIFs蛋白结构信息
Table 5 Structural information of FmPIFs protein in F. mandshurica
蛋白名称
Protein
nameα螺旋占比
Alpha
helix proportion/%延伸链占比
Extended
strand proportion/%无规则卷曲占比
Random
coil proportion/%保守结构域位点
Conserved domain
site亲水区/疏水区数目
Number of hydrophilic/
hydrophobic zone定位预测及得分
Position prediction
and scoreFmPIF1 25.48 18.92 55.60 310 ~ 373 296/32 细胞核 Nucleus 14 FmPIF3 18.01 14.71 67.19 444 ~ 506 366/66 细胞核 Nucleus 13 FmPIF4 26.09 19.22 54.69 253 ~ 316 272/22 细胞核 Nucleus 13 FmPIF7 36.49 10.39 53.12 242 ~ 305 224/51 细胞核 Nucleus 11 FmPIF8 21.41 25.06 53.53 239 ~ 302 220/75 细胞核 Nucleus 14 -
[1] 李秀坤, 许冬清. 植物光信号转导[J]. 自然杂志, 2019, 41(3):183−187. doi: 10.3969/j.issn.0253-9608.2019.03.004 Li X K, Xu D Q. Plant light signal transduction[J]. Journal of Nature, 2019, 41(3): 183−187. doi: 10.3969/j.issn.0253-9608.2019.03.004
[2] Gabriela T O. The Arabidopsis basic/helix-loop-helix transcription factor family[J]. The Plant Cell, 2003, 8(15): 1749−1770.
[3] Yu Z. A quartet of pif bhlh factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis [J/OL]. PLoS Genetics, 2013, 1(9): e1003244[2020−10−31]. https://doi.org/10.1371/journal.pgen.1003244.
[4] Liu X, Chen C Y, Wang K C, et al. Phytochrome interacting factor3 associates with the histone deacetylase hda15 in repression of chlorophyll biosynthesis and photosynthesis in etiolated Arabidopsis seedlings[J]. The Plant Cell, 2013, 25(4): 1258−1273. doi: 10.1105/tpc.113.109710
[5] Leivar P, Monte E, Oka Y, et al. Multiple phytochrome-interacting bhlh transcription factors repress premature seedling photomorphogenesis in darkness[J]. Current Biology, 2008, 18(23): 1815−1823. doi: 10.1016/j.cub.2008.10.058
[6] Leivar P, Quail P H. Pifs: pivotal components in a cellular signaling hub[J]. Trends in Plant Science, 2011, 16(1): 19−28. doi: 10.1016/j.tplants.2010.08.003
[7] 杨剑飞, 王宇, 杨琳, 等. 光敏色素互作因子PIFs是整合多种信号调控植物生长发育的核心元件[J]. 植物生理学报, 2014, 50(8):1109−1118. Yang J F, Wang Y, Yang L, et al. Phytochrome interaction factors pifs are the core elements that integrate multiple signals to regulate plant growth and development[J]. Acta Plant Physiology, 2014, 50(8): 1109−1118.
[8] 任小芸. ZmPIFs基因的克隆、表达及AtPIFs基因的抗旱功能研究[D]. 扬州: 扬州大学, 2017. Ren X Y. Cloning and expression of ZmPIFs gene and research on drought resistance function of AtPIFs gene[D]. Yangzhou: Yangzhou University, 2017.
[9] Wang F, Chen X, Dong S, et al. Crosstalk of PIF4 and DELLA modulates CBF transcript and hormone homeostasis in cold response in tomato[J]. Plant Biotechnology Journal, 2020, 18(4): 1041−1055. doi: 10.1111/pbi.13272
[10] Lau O S, Deng X W. Plant hormone signaling lightens up: integrators of light and hormones[J]. Current Opinion in Plant Biology, 2010, 13(5): 571−577. doi: 10.1016/j.pbi.2010.07.001
[11] Nozue K, Harmer S L, Maloof J N. Genomic analysis of circadian clock-, light-, and growth-correlated genes reveals phytochrome-interacting factor 5 as a modulator of auxin signaling in Arabidopsis[J]. Plant Physiology, 2011, 156(1): 357−372. doi: 10.1104/pp.111.172684
[12] Feng S H, Cristina M, Giuliana G, et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins[J]. Nature, 2008, 451: 475−479.
[13] Richter R, Behringer C, Muller I K, et al. The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS[J]. Genes & Development, 2010, 24(18): 2093−2104.
[14] Keara A F, Sang H L, Dhaval P, et al. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature[J]. PNAS, 2011, 108(50): 20231−20235. doi: 10.1073/pnas.1110682108
[15] Friml J Í, Wisniewska J, Benková E, et al. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis[J]. Nature, 2002, 415: 806−809. doi: 10.1038/415806a
[16] Sun J Q, Qi L L, Li Y N, et al. Pif4-mediated activation of yucca8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. [J/OL]. PLoS Genetics, 2012, 8(3): e1002594[2020−09−20]. https://doi.org/10.1371/journal.pgen.1002594.
[17] Leivar P, Tepperman J M, Cohn M M, et al. Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis[J]. The Plant Cell, 2012, 24(4): 1398−1419. doi: 10.1105/tpc.112.095711
[18] Leivar P, Monte E. PIFs: systems integrators in plant development[J]. The Plant Cell, 2014, 26(1): 56−78. doi: 10.1105/tpc.113.120857
[19] Bernardo-García S, Delucas M, Martínez C, et al. BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth.[J]. Genes & Development, 2014, 28(15): 1681−1694.
[20] Kim J, Kang H, Park J, et al. PIF1-interacting transcription factors and their binding sequence elements determine the in vivo targeting sites of PIF1[J]. The Plant Cell, 2016, 28(6): 1388−1405. doi: 10.1105/tpc.16.00125
[21] 周明琦. CBF信号途径在低温下对植物生长的调控及其育种应用[D]. 上海: 复旦大学, 2013. Zhou M Q. Regulation of CBF signaling pathway on plant growth under low temperature and its breeding application [D]. Shanghai: Fudan University, 2013.
[22] Jiang B, Shi Y, Zhang X, et al. PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis[J]. PNAS, 2017, 114(32): E6695−E6702. doi: 10.1073/pnas.1706226114
[23] 王峰. PhyA、HY5和PIF4在光质调控番茄低温抗性中的机制研究[D]. 杭州: 浙江大学, 2017. Wang F. Mechanism of Phya, HY5 and PIF4 in light quality regulation of low temperature resistance in tomato [D]. Hangzhou: Zhejiang University, 2017.
[24] Gao Y, Jiang W, Dai Y, et al. A maize phytochrome-interacting factor 3 improves drought and salt stress tolerance in rice[J]. Plant Molecular Biology, 2015, 87(4/5): 413−428.
[25] Kumar S V, Lucyshyn D, Jaeqer K E, et al. Transcription factor PIF4 controls the thermosensory activation of flowering [J]. Nature, 2012, 484: 242−245.
[26] 赵晓玲. 植物中与光敏色素相互作用的因子PIFs[J]. 植物生理学通讯, 2009, 45(6):531−536. Zhao X L. PIFs interacting with phytochrome in plants[J]. Journal of Plant Physiology, 2009, 45(6): 531−536.
[27] Soy J, Leivar P, Nahuel G, et al. Phytochrome-imposed oscillations in PIF3 protein abundance regulate hypocotyl growth under diurnal light/dark conditions in Arabidopsis[J]. The Plant Journal, 2012, 71(3): 390−401.
[28] Herbert K, Hans-Joachim G. The protein protocols handbook, 2nd edition [M]. Totowa: Humana Press, 2002.
[29] 孙平楠, 周小玲, 王正祥. 信号肽生物信息学分析在Neurospora crassa phyA基因鉴定中的应用[J]. 南方医科大学学报, 2009, 29(6):1098−1101. doi: 10.3321/j.issn:1673-4254.2009.06.003 Sun P N, Zhou X L, Wang Z X. Application of signal peptide bioinformatics analysis in identification of Neurospora crassa phyA gene[J]. Journal of Southern Medical University, 2009, 29(6): 1098−1101. doi: 10.3321/j.issn:1673-4254.2009.06.003
[30] Thomas N P, Soren B, Gunnar V H, et al. Signalp 4.0: discriminating signal peptides from transmembrane regions[J]. Nature Methods, 2011, 8(10): 785−786. doi: 10.1038/nmeth.1701
[31] Horton P, Park K J, Obayashi T, et al. Wolf psort: protein localization predictor [J]. Nucleic Acids Research, 2007, 35(Suppl.2): W585−W587.
[32] Hu B, Jin L, Guo A Y, et al. GSDS 2.0: an upgraded gene feature visualization server[J]. Bioinformatics, 2015, 31(8): 1296−1297. doi: 10.1093/bioinformatics/btu817
[33] Ma B, Yuan Y, Gao M, et al. Genome-wide identification, molecular evolution, and expression divergence of aluminum-activated malate transporters in apples[J]. International Journal of Molecular Sciences, 2018, 19(9): 2807−2809. doi: 10.3390/ijms19092807
[34] Larkin M. Clustal W and Clustal X version 2.0[J]. Bioinformatics, 2007, 23(21): 2947−2948. doi: 10.1093/bioinformatics/btm404
[35] Olsson H, Belfrage P. Phosphorylation and dephosphorylation of hormone-sensitive lipase interactions between the regulatory and basal phosphorylation sites[J]. FEBS Letters, 1988, 232(1): 78−82.
[36] 任小龙, 詹亚光, 梁雪, 等. 水曲柳花发育过程中AG、SOC1基因表达的qRT-PCR分析[J]. 植物研究, 2015, 35(4):612−617. doi: 10.7525/j.issn.1673-5102.2015.04.021 Ren X L, Zhan Y G, Liang X, et al. QRT-PCR analysis of AG and SOC1 gene expression during flower development of Fraxinus mandshurica[J]. Plant Research, 2015, 35(4): 612−617. doi: 10.7525/j.issn.1673-5102.2015.04.021
[37] 韦雪芳, 王冬梅, 刘思, 等. 信号肽及其在蛋白质表达中的应用[J]. 生物技术通报, 2006(6):38−42. doi: 10.3969/j.issn.1002-5464.2006.06.009 Wei X F, Wang D M, Liu S, et al. Signal peptide and its application in protein expression[J]. Biotechnology Bulletin, 2006(6): 38−42. doi: 10.3969/j.issn.1002-5464.2006.06.009
[38] 王镜岩, 朱圣庚, 徐长法. 生物化学[M]. 北京: 高等教育出版社, 2002. Wang J Y, Zhu S G, Xu C F. Biochemistry[M]. Beijing: Higher Education Press, 2002.
[39] 宋毓峰. 林烟草钾转运体基因NsHAK11的克隆与功能分析[D]. 北京: 中国农业科学院, 2014. Song Y F. Cloning and functional analysis of tobacco potassium transporter gene NsHAK11 [D]. Beijing: Chinese Academy of Agricultural Sciences, 2014.
[40] 高凯. NJ进化树构建方法的改进及其应用[D]. 北京: 北京工业大学, 2008. Gao K. Improvement and application of NJ evolutionary tree construction method [D]. Beijing: Beijing University of Technology, 2008.
[41] Jeong J, Choi G. Phytochrome-interacting factors have both shared and distinct biological roles[J]. Molecules and Cells, 2013, 35(5): 371−380. doi: 10.1007/s10059-013-0135-5
[42] Alabadí D, Gallego-Bartolomé J, Orlando L, et al. Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness[J]. Plant Journal, 2010, 53(2): 324−335.
[43] Shen H, Zhu L, Castillon A, et al. Light-induced phosphorylation and degradation of the negative regulator PIF1 depends upon its direct physical interactions with photoactivated phytochromes[J]. The Plant Cell, 2008, 20(6): 1586−1602. doi: 10.1105/tpc.108.060020
[44] 宋晓祎. 玉米中ZmPIFs基因的克隆与功能分析[D]. 泰安: 山东农业大学, 2016. Song X Y. Cloning and functional analysis of ZmPIFs gene in Zea mays [D]. Taian: Shandong Agricultural University, 2016.
[45] 刘浩浩. 过表达ZmPIF3转基因拟南芥植株抗盐分析[D]. 郑州: 河南农业大学, 2018. Liu H H. Salt tolerance analysis of transgenic Arabidopsis plants overexpressing ZmPIF3 [D]. Zhengzhou: Henan Agricultural University, 2018.
[46] 刘春浩, 梁楠松, 于磊, 等. 水曲柳TCP4转录因子克隆及胁迫和激素下的表达分析[J]. 北京林业大学学报, 2017, 39(6):22−31. Liu C H, Liang N S, Yu L, et al. Cloning and expression analysis of TCP4 transcription factor in Fraxinus mandshurica[J]. Journal of Beijing Forestry University, 2017, 39(6): 22−31.
[47] Zheng P F, Wang X, Yang Y Y, et al. Identification of phytochrome-interacting factor family members and functional analysis of MdPIF4 in Malus domestica [J/OL]. International Journal of Molecular Sciences, 2020, 21(19): 7350[2020−11−11] . https://doi.org/10.3390/ijms21197350.
[48] 潘教文, 赵术珍, 张烨, 等. 光敏色素互作因子(PIFs)对植物生长发育的调控[J]. 山东农业科学, 2014, 46(6):150−156. Pan J W, Zhao S Z, Zhang Y, et al. Regulation of phytochrome interaction factors (PIFs) on plant growth and development[J]. Shandong Agricultural Sciences, 2014, 46(6): 150−156.
[49] Oh E, Kang H, Yamaguchi S, et al. Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE 5 during seed germination in Arabidopsis[J]. The Plant Cell, 2009, 21(2): 403−419. doi: 10.1105/tpc.108.064691



 下载:
下载: