Development and application of EST-SSR molecular markers in Pinus bungeana
-
摘要:目的 白皮松作为我国濒危的乡土树种,具有重要的经济和园林观赏价值,为进一步开发和利用白皮松种质资源,本研究基于EST-SSR标记对北京地区3个不同种源地的白皮松群体遗传多样性开展评价。方法 以白皮松转录组数据为依据,对其微卫星位点进行筛选并设计合成了96对引物,并对北京温泉苗圃栽培收集的3个不同种源地(北京、山东、山西)的白皮松群体,共60个植株开展遗传多样性分析,对其群体内与群体间的遗传结构进行评价。结果 96对引物中共筛选获得了5对多态性引物,其观察杂合度、期望杂合度、多态性信息含量和等位基因位点个数变化范围分别为0.203 ~ 0.433、0.211 ~ 0.530、0.187 ~ 0.484和2 ~ 5。3个白皮松样本群体中等位基因数量、有效等位基因数量、香农信息指数、观察杂合度和固定指数变化范围为2.400 ~ 3.000、1.516 ~ 1.761、0.484 ~ 0.606、0.295 ~ 0.362和−0.075 ~ 0.081,平均值分别为2.677、1.632、0.560、0.333和−0.007。遗传分化系数和基因流变化范围分别为0.021 6 ~ 0.115 3和1.399 6 ~ 11.340 0,平均值分别为0.090 2和2.521 2。AMOVA分析显示遗传变异主要来自于群体内,群体间差异较小,仅占据11%。结论 本研究共获得了5对多态性的白皮松EST-SSR引物,可用于后续白皮松群体遗传多样性分析和分子标记辅助育种工作;北京温泉苗圃栽培收集的3个不同种源地的白皮松群体分析结果表明,当地现有收集的白皮松种源群体遗传相似度较高,在未来种质资源保存和苗木繁育工作中应考虑增加其他种源地的白皮松群体。Abstract:Objective As an endangered native tree species in China, Pinus bungeana has important economic and garden ornamental values. In order to further develop and utilize the species germplasm resources, the study evaluated the genetic diversity of three populations from different provenances in P. bungeana.Method Simple sequence repeats were selected from the transcriptome data in P. bungeana, and then 96 pairs of primers were developed, and the genetic diversity of 60 individuals in Beijing Hot Spring Nursery collected from three different provenances (Beijing, Shandong, and Shanxi) was analyzed, and genetic diversity parameters between intra- and inter-population were analyzed.Result The variation ranges of the observed heterozygosity, expected heterozygosity, polymorphism information content and the number of allele loci for 5 pairs of polymorphic primers were 0.203−0.433, 0.211−0.530, 0.187−0.484 and 2−5, respectively. The number of alleles, effective alleles, Shannon diversity index, observed heterozygosity and fixed index of the three populations ranged in 2.400−3.000, 1.516−1.761, 0.484−0.606, 0.295−0.362 and −0.075−0.081, the average values were 2.677, 1.632, 0.560, 0.333 and −0.007, respectively. The ranges of genetic differentiation coefficient and gene flows were 0.021 6−0.115 3 and 1.399 6−11.340 0, and the average values were 0.090 2 and 2.521 2, respectively. AMOVA analysis showed that genetic variation mainly comed from within the population, with small differences between populations, accounting for only 11%.Conclusion This study obtained 5 pairs of polymorphic EST-SSR primers in P. bungeana, which can be used for subsequent analysis of the population genetic diversity and molecular marker-assisted breeding. Genetic diversity analysis about the three populations of P. bungeana shows that the existing populations in Beijing Hot Spring Nursery have high genetic similarity, and the populations from other provenances should be considered for the future germplasm resource preservation and seedling breeding work.
-
油橄榄(Olea europaea)主要分布于地中海地区,是一种重要的木本油料作物。橄榄油中富含健康的不饱和脂肪酸、酚类化合物和维生素E等营养物质,不仅是地中海式饮食的重要成分,而且远销美国、墨西哥、巴西、印度、澳大利亚等国家[1]。近年来,全球橄榄油需求量增加,产量却十分有限。要持续扩大油橄榄的适生区,增加橄榄油的产量,种质选育创制尤为关键。随着测序技术的飞速发展,油橄榄已完成了高质量基因组文件的组装和注释[2],并获得了丰富的转录组数据,这为我们借助分子育种选育抗逆性强、产油率高的油橄榄奠定了基础。
过氧化物酶(peroxidase,PRX)是一类广泛存在于生物体内的酶,它通过将过氧化氢还原为水来催化底物的氧化。根据其蛋白序列和结构特征,PRX可分为2类,血红素类和非血红素类;血红素PRX又可进一步分为动物型和非动物型;非动物的PRX包含3个小类,Ⅰ、Ⅱ和Ⅲ类,这3小类PRX具有非常相似的三维结构,但氨基酸序列同源性较低,且具有不同的功能和反应机制[3]。Ⅰ类PRX广泛分布于非动物性生命体内,其在原核生物中的分布表明其他2类PRX可能起源于Ⅰ类PRXs[4],Ⅰ类PRX包含抗坏血酸PRX、细胞色素c PRX和过氧化氢酶,主要功能是消除过量的H2O2[5-6]。Ⅱ类PRX仅分布于真菌中,包含锰PRX、木质素PRX和其他PRX[7],主要功能是降解木质素。Ⅲ类PRX分布于陆生植物体内,是一个庞大的多基因家族,通常分泌进入细胞壁或其周围基质或液泡中。Ⅲ类PRX广泛参与植物生命周期内的一系列的生理活动[8],比如细胞壁形成[9]、伤口愈合[10]、生长素代谢[11-12]等。另外,Ⅲ类PRX通过调节多种活性氧水平响应生物胁迫和非生物胁迫[12],因此,研究Ⅲ类PRX基因功能对作物遗传育种具有十分重要的作用。
目前,拟南芥(Arabidopsis thaliana)[13]、水稻(Oryza sativa)[14]、玉米(Zea mays)[15]、毛果杨(Populus trichocarpa)[16]、白梨(Pyrus bretschneideri)[17]、葡萄(Vitis vinifera)[18]、木薯(Manihot esculenta)[19]和小麦(Triticum aestivum)[20]等植物的Ⅲ类PRX基因全基因组鉴定和分析已经完成,这些物种中有47 ~ 138个不等的Ⅲ类PRX基因。单个Ⅲ类PRX基因功能的研究也日益丰富,例如,AtPRX4、AtPRX52和AtPRX72拟南芥突变体木质素含量降低,木质素组成发生改变[21-23]。烟草(Nicotiana tabacum)中过表达AtPRX64促进根系生长,减少活性氧和铝在根系中的积累,从而提高对铝胁迫的耐受性[24];拟南芥中过表达OsPRX38促进胼胝质沉积、抗氧化酶活性和叶绿素含量上升,从而提高植物对砷胁迫的耐受性[25]。然而,关于油橄榄Ⅲ类PRX基因家族的全基因组鉴定及基因功能研究仍未见报道。本研究利用最近组装注释的油橄榄基因组和公共数据库中的转录组信息,完成了油橄榄Ⅲ类PRX家族的鉴定、进化和表达分析。研究发现:油橄榄是目前已知Ⅲ类PRX家族成员最多的双子叶植物,其复制基因的亚功能化和新功能化对油橄榄在地中海地区的广泛分布具有十分重要的意义。
1. 材料与方法
1.1 油橄榄Ⅲ类PRX基因家族成员的鉴定
油橄榄基因组文件下载于NGDC(https://bigd.big.ac.cn/search/?dbId=gwh&q=Olea%20europaea&page=1),Ⅲ类PRX基因家族的保守结构域(PF00141)的HMM模型下载于pfam(http://pfam.xfam.org/family/PF00141#tabview=tab6)。利用PF00141的HMM模型,在油橄榄蛋白质注释文件(GWHAOPM00000000.Protein.faa)中进行HMM模型搜索(hmmsearch),得到120条蛋白序列。利用拟南芥的73条PRX蛋白序列[13]在油橄榄基因组文件(GWHAOPM00000000.Protein.faa)中进行本地化搜索(blastp),E值设置为10−6,得到119条序列。2种方法得到的共有序列为119条。将119条序列上传至Pfam(http://pfam.xfam.org/search#tabview=tab1)和NCBI-CDD(https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi)数据库中,预测其是否含有Ⅲ类PRX基因家族的保守结构域从而进行筛选。Pfam鉴定结果表明,119条序列均含有PF00141结构域;NCBI-CDD鉴定结果表明,13条序列含有不完整的分泌类过氧化物酶保守结构域(secretory_peroxidase,cd00693)。因此,最后选定106条序列作为油橄榄Ⅲ类PRX基因成员。利用Prot-Param(https://web.expasy.org/compute_pi/)预测等电点和分子量;利用SignalP-5.0 Server(http://www.cbs.dtu.dk/services/SignalP/)预测信号肽Sec/SPI的存在概率及长度,Signal peptide likelihood值大于0.5的序列视为含有信号肽,反之认为不含有信号肽;利用Wolfpsort(https://wolfpsort.hgc.jp/)预测亚细胞定位。
1.2 PRX蛋白系统进化树的构建
利用TBtools v1.095中的Fasta Extract从油橄榄基因组文件中提取得到106个OePRX的蛋白序列,从前人的论文中下载得到拟南芥和毛果杨PRX蛋白序列[16],其中,拟南芥73条,毛果杨93条。利用MEGA7.0中的ClustalW比对272条全长的PRX蛋白序列,并使用邻接法构建系统进化树,设置替代模型为Poisson model,设置缺失数据处理方式为Pairwise deletion,设置bootstrap值为1 000。
1.3 OePRX染色体定位和复制基因分析
利用TBtools v1.095中的Fasta Stats从油橄榄基因组文件中获取23条染色体的长度和OePRX基因家族成员的位置信息,利用MapChart绘制OePRX基因家族在23条染色体上的分布,需注意的是,OePRX基因家族成员中100个基因分布于染色体上,但6个基因分布于Contig上,这6个基因未在图中展示。利用TBtools v1.095中的One Step MCScanX分析OePRX基因家族中的复制基因;利用MEGA7.0中的Muscle (Codons)比对复制基因对的编码区(CDS),再利用KaKs_Calculator2.0计算复制基因对的非同义替换(Ka)和同义替换(Ks)值,设置计算方式为YN。拟南芥基因组文件下载自http://ensembl.gramene.org/Arabidopsis_thaliana/Info/Index,毛果杨基因组文件下载自https://phytozome.jgi.doe.gov。利用同样的方法分析油橄榄和拟南芥及毛果杨PRX之间的复制基因,并计算Ka和Ks值。利用TBtools v1.095中的Advanced Circos绘制油橄榄PRX基因线性图;用Multiple Synteny Plot绘制油橄榄与拟南芥和毛果杨PRX基因的线性图。
1.4 OePRX基因序列分析
利用TBtools v1.095中的Fasta Extract从油橄榄基因组文件提取OePRX基因家族成员的DNA序列和CDS序列,再利用GSDS2.0(http://gsds.gao-lab.org/)绘制其基因结构图。利用在线网站MEME(https://meme-suite.org)预测OePRX基因的保守基序,预测基序的个数设置为10,其他参数默认。利用TBtools v1.095中的Fasta Extract从油橄榄基因组文件中获取106个OePRX基因上游1 500 bp的序列作为启动子,并将其提交至PlantCARE(http://bioinformatics.psb.ugent.be)预测启动子上的顺式作用元件,最后对结果进行分析整理。
1.5 OePRX基因表达模式分析
利用NCBI(https://www.ncbi.nlm.nih.gov/)中油橄榄不同组织[26](根、茎、叶、花、果和分生组织)、油橄榄枝条响应干热胁迫及复水[27]和油橄榄叶片响应水涝胁迫的RNA-seq数据(未发表)分析OePRX基因家族的表达模式。使用sra explorer获取测序数据的FTP地址,使用fastp去除接头和低质量的reads,利用hisat2比对并使用htseq-count获取基因的原始的读长数目(reads counts),使用DEseq2进行差异表达分析。自行编写R脚本将原始读长数目转换为tpm(每千个碱基的转录每百万映射读取的转录本,均一化后的read counts值)。差异基因鉴定标准为FDR ≤ 0.05,并且|log2(Fold Change)| ≥ 2(Fold Change为差异倍数)。取log2(tpm + 1),利用R语言绘制热图。
1.6 实时荧光定量PCR
油橄榄品种“科罗莱卡”种植于中国林科院林木遗传育种国家重点实验室温室,选择3株长势均一的3年生油橄榄幼苗,剪取叶片、茎、根和果实,于液氮中带回实验室后冻存于−80 ℃冰箱内。利用MiniBEST植物RNA提取试剂盒(TaKaRa,9769)提取不同组织的总RNA;利用PrimeScript™ RT Master Mix(Takara,RR036A)反转录合成cDNA;利用Primer Premier5.0设计引物;利用TB Green® Premix Ex Taq™ Ⅱ(Tli RNaseH Plus)(Takara,RR820A)在Roche Light Cycler 480上进行实时荧光定量PCR(qRT-PCR),反应体系如下:总体系20 μL,包括TB Green Premix Ex Taq Ⅱ(Tli RNaseH Plus)(2×)10 μL,cDNA 2 μL,正、反向引物各0.8 μL,去离子水6.4 μL。反应程序为2步法,详见试剂盒说明书。以Actin[28]为内参基因,利用2−△△CT法计算基因相对表达量。
2. 结果与分析
2.1 OePRX基因家族成员的鉴定
血红素过氧化物酶(haem peroxidase)的保守结构域是PF00141,这类酶大都含有2个α结构域,中间嵌有血红素基团[29]。利用PF00141的HMM模型和73条AtPRX蛋白在油橄榄蛋白质序列文件中分别进行hmmsearch和blastp,将2种方法得到的结果取交集,得到了119条候选PRX。植物Ⅲ类过氧化物酶特有的结构域是分泌类过氧化物酶保守结构域(secretory_peroxidase,cd00693),这类酶含有4个保守的二硫桥和2个保守的钙离子结合位点[30]。根据NCBI-CDD预测结果,删除分泌类过氧化物酶保守结构域不完整的13条序列,最后得到了106条油橄榄Ⅲ类PRX蛋白序列(表1)。根据其在染色体上的分布情况,将其命名为OePRX1-OePRX106。OePRX蛋白的氨基酸个数最小为240,最大为509;分子量最小为26.55 kDa,最大为58.00 kDa;等电点变化范围为4.45 ~ 9.75,其中74个OePRX蛋白(69.8%)大于等于7。通过在线预测发现:75个OePRX蛋白(70.8%)含有Sec/SPI信号肽,信号肽长度变化范围为19 ~ 32个氨基酸;值得注意的是,组14中的10个OePRX蛋白均不含有信号肽。亚细胞定位预测结果显示:含有信号肽的OePRX蛋白大多位于叶绿体、质膜、液泡膜和内质网(50个OePRX蛋白,占含有信号肽的OePRX蛋白总数的66.7%),也有一部分位于细胞外和细胞质(25个OeORXs蛋白,占含有信号肽的OePRX总数的33.3%);不含信号肽的OePRX蛋白(31个OePRX蛋白)中,11个OePRX蛋白位于细胞外和细胞质,其余位于叶绿体、内质网、细胞核和质膜。
表 1 油橄榄Ⅲ类PRX 基因的命名及其理化性质Table 1. Nomenclature and physicochemical properties of class Ⅲ PRX genes from olive名称 Name ID 内含子相位
Intron phase蛋白质长度
Protein length/aa分子量
Molecular mass/kDa等电点
Isoelectric point信号肽
Signal peptide预测的亚细胞定位
Predicted subcellular localizationOePRX1 EVM0022477 0 344 38.40 6.10 无 No 胞外 Extracellular OePRX2 EVM0032926 0 266 29.38 8.97 无 No 细胞质 Cytosol OePRX3 EVM0020804 0 327 35.65 9.21 有 Yes 叶绿体 Chloroplast OePRX4 EVM0047797 100 327 35.92 8.24 无 No 细胞核 Nucleus OePRX5 EVM0009292 100 316 34.40 8.72 有 Yes 液泡膜 Vacular membrane OePRX6 EVM0060238 0 330 35.97 8.89 无 No 叶绿体 Chloroplast OePRX7 EVM0022620 12001000 276 30.39 8.66 无 No 细胞质 Cytosol OePRX8 EVM0017319 1200201 279 30.58 7.70 无 No 细胞质 Cytosol OePRX9 EVM0047458 20010 250 27.43 5.67 无 No 细胞质 Cytosol OePRX10 EVM0057886 0 319 34.48 6.98 有 Yes 叶绿体 Chloroplast OePRX11 EVM0047784 0 339 37.00 7.48 有 Yes 质膜 Plasma membrane OePRX12 EVM0039807 20 358 38.84 7.51 有 Yes 叶绿体 Chloroplast OePRX13 EVM0011450 0 317 34.35 9.14 有 Yes 叶绿体 Chloroplast OePRX14 EVM0019223 1 309 35.27 9.41 有 Yes 胞外 Extracellular OePRX15 EVM0012728 1 320 36.91 9.20 有 Yes 细胞质 Cytosol OePRX16 EVM0000231 0 336 37.28 7.08 有 Yes 叶绿体 Chloroplast OePRX17 EVM0024793 0 322 35.21 8.38 有 Yes 叶绿体 Chloroplast OePRX18 EVM0042519 0 321 34.96 8.76 有 Yes 叶绿体 Chloroplast OePRX19 EVM0059982 0 321 34.90 8.77 有 Yes 叶绿体 Chloroplast OePRX20 EVM0019685 0 333 36.73 5.68 无 No 质膜 Plasma membrane OePRX21 EVM0058087 0 324 35.74 8.53 有 Yes 细胞质 Cytosol OePRX22 EVM0027348 0 323 35.02 5.82 有 Yes 液液泡膜 Vacular membrane OePRX23 EVM0060830 0 331 36.34 9.41 有 Yes 胞外 Extracellular OePRX24 EVM0015621 0 247 27.39 8.88 无 No 细胞核 Nucleus OePRX25 EVM0010445 12101221 327 35.56 9.26 无 No 叶绿体 Chloroplast OePRX26 EVM0006524 0 338 37.53 6.45 有 Yes 胞外 Extracellular OePRX27 EVM0042768 22 304 32.63 6.58 有 Yes 叶绿体 Chloroplast OePRX28 EVM0048359 0 355 37.59 8.35 有 Yes 叶绿体 Chloroplast OePRX29 EVM0028664 0 317 34.49 9.09 有 Yes 液泡膜 Vacular membrane OePRX30 EVM0035621 0 327 34.85 5.51 有 Yes 胞外 Extracellular OePRX31 EVM0048816 100 324 36.67 6.26 有 Yes 胞外 Extracellular OePRX32 EVM0003637 1002 350 39.07 8.83 有 Yes 胞外 Extracellular OePRX33 EVM0019330 0 333 36.44 9.48 有 Yes 液泡膜 Vacular membrane OePRX34 EVM0033281 2100102 273 30.15 7.73 无 No 细胞质 Cytosol OePRX35 EVM0015060 10100000 319 35.73 6.63 无 No 叶绿体 Chloroplast OePRX36 EVM0017946 0 329 37.29 8.48 有 Yes 细胞质 Cytosol OePRX37 EVM0021366 − 331 36.79 9.34 有 Yes 叶绿体 Chloroplast OePRX38 EVM0060527 0 342 37.64 6.21 有 Yes 细胞质 Cytosol OePRX39 EVM0014494 0 321 35.37 6.18 有 Yes 胞外 Extracellular OePRX40 EVM0042196 0 319 34.12 9.41 有 Yes 叶绿体 Chloroplast OePRX41 EVM0015681 0 346 38.94 8.86 有 Yes 内质网 Endoplasmic reticulum OePRX42 EVM0047610 0 327 35.27 8.80 有 Yes 叶绿体 Chloroplast OePRX43 EVM0047452 20002 379 41.95 8.87 无 No 叶绿体 Chloroplast OePRX44 EVM0061929 0 319 34.23 4.45 有 Yes 胞外 Extracellular OePRX45 EVM0054999 0 291 32.27 5.44 无 No 胞外 Extracellular OePRX46 EVM0048606 0 328 35.67 6.51 有 Yes 液泡膜 Vacular membrane OePRX47 EVM0037058 20 315 34.41 7.55 无 No 内质网 Endoplasmic reticulum OePRX48 EVM0022847 20 413 45.85 9.56 无 No 叶绿体 Chloroplast OePRX49 EVM0020385 20 373 41.16 9.35 无 No 叶绿体 Chloroplast OePRX50 EVM0045080 0 317 34.55 9.02 有 Yes 叶绿体 Chloroplast OePRX51 EVM0044622 0 320 35.53 9.54 有 Yes 叶绿体 Chloroplast OePRX52 EVM0021425 0 324 36.14 6.17 有 Yes 胞外 Extracellular OePRX53 EVM0045494 0 327 35.52 8.71 有 Yes 液泡膜 Vacular membrane OePRX54 EVM0020915 20 306 33.46 8.90 有 Yes 液泡膜 Vacular membrane OePRX55 EVM0031491 0 314 34.07 9.32 有 Yes 叶绿体 Chloroplast OePRX56 EVM0005944 2 292 31.79 9.45 有 Yes 叶绿体 Chloroplast OePRX57 EVM0015088 0 317 34.42 9.13 有 Yes 胞外 Extracellular OePRX58 EVM0050632 0 314 34.04 9.25 有 Yes 叶绿体 Chloroplast OePRX59 EVM0028088 0 317 34.23 8.99 有 Yes 胞外 Extracellular OePRX60 EVM0025336 0 326 34.42 7.58 无 No 叶绿体 Chloroplast OePRX61 EVM0012469 0 317 34.68 9.33 有 Yes 胞外 Extracellular OePRX62 EVM0049574 0 340 37.08 6.25 有 Yes 胞外 Extracellular OePRX63 EVM0050273 10 240 26.55 6.22 无 No 叶绿体 Chloroplast OePRX64 EVM0050817 0 310 34.15 8.46 有 Yes 叶绿体 Chloroplast OePRX65 EVM0009492 0 322 34.85 4.99 有 Yes 叶绿体 Chloroplast OePRX66 EVM0047271 1200201 333 36.96 8.64 无 No 质膜 Plasma membrane OePRX67 EVM0005889 210122 509 58.00 8.73 无 No 细胞核 Nucleus OePRX68 EVM0026529 0 331 36.43 9.50 有 Yes 液泡膜 Vacular membrane OePRX69 EVM0038965 20 310 33.90 9.23 有 Yes 质膜 Plasma membrane OePRX70 EVM0024841 0 328 35.72 8.37 有 Yes 叶绿体 Chloroplast OePRX71 EVM0047959 3 314 34.07 6.94 有 Yes 叶绿体 Chloroplast OePRX72 EVM0059042 0 339 37.43 5.56 有 Yes 质膜 Plasma membrane OePRX73 EVM0022158 0 318 34.18 6.88 有 Yes 叶绿体 Chloroplast OePRX74 EVM0027175 0 326 36.14 9.75 有 Yes 叶绿体 Chloroplast OePRX75 EVM0057531 20 287 30.73 8.89 有 Yes 液泡膜 Vacular membrane OePRX76 EVM0032201 0 293 32.34 8.75 无 No 叶绿体 Chloroplast OePRX77 EVM0059919 0 339 37.61 8.36 有 Yes 液泡膜 Vacular membrane OePRX78 EVM0011789 0 355 39.51 8.06 无 No 质膜 Plasma membrane OePRX79 EVM0024970 − 329 36.54 9.19 有 Yes 叶绿体 Chloroplast OePRX80 EVM0047604 100 339 37.82 8.30 无 No 细胞质 Cytosol OePRX81 EVM0061300 0 330 36.94 5.94 有 Yes 叶绿体 Chloroplast OePRX82 EVM0042525 0 298 32.64 8.62 有 Yes 胞外 Extracellular OePRX83 EVM0045066 0 318 34.03 9.15 有 Yes 叶绿体 Chloroplast OePRX84 EVM0029808 0 354 39.38 5.68 无 No 胞外 Extracellular OePRX85 EVM0032932 1022010 247 27.30 5.65 无 No 细胞质 Cytosol OePRX86 EVM0038819 2001020 250 27.72 5.44 无 No 细胞质 Cytosol OePRX87 EVM0002745 200 321 34.58 4.77 无 No 细胞核 Nucleus OePRX88 EVM0038873 200 326 34.71 8.30 有 Yes 液泡膜 Vacular membrane OePRX89 EVM0026260 2 348 38.22 9.56 有 Yes 质膜 Plasma membrane OePRX90 EVM0044542 0 296 32.61 8.14 有 Yes 胞外 Extracellular OePRX91 EVM0046395 0 318 34.24 9.54 有 Yes 叶绿体 Chloroplast OePRX92 EVM0042595 0 316 33.95 7.51 有 Yes 叶绿体 Chloroplast OePRX93 EVM0047351 0 320 35.08 5.48 有 Yes 叶绿体 Chloroplast OePRX94 EVM0045920 0 332 35.75 4.63 有 Yes 胞外 Extracellular OePRX95 EVM0044144 0 325 35.63 8.75 无 No 叶绿体 Chloroplast OePRX96 EVM0048820 0 314 35.88 8.61 有 Yes 细胞质 Cytosol OePRX97 EVM0058650 0 350 38.67 6.94 有 Yes 叶绿体 Chloroplast OePRX98 EVM0026875 0 371 40.68 6.31 无 No 叶绿体 Chloroplast OePRX99 EVM0019820 0 340 36.88 6.59 有 Yes 胞外 Extracellular OePRX100 EVM0018143 20020200000 394 43.59 9.28 无 No 叶绿体 Chloroplast OePRX101 EVM0029423 0 343 37.01 8.94 有 Yes 叶绿体 Chloroplast OePRX102 EVM0036633 0 342 37.59 9.17 有 Yes 胞外 Extracellular OePRX103 EVM0041887 1 312 33.33 5.97 有 Yes 叶绿体 Chloroplast OePRX104 EVM0032297 0 327 35.71 8.57 有 Yes 胞外 Extracellular OePRX105 EVM0042746 0 302 33.53 8.96 有 Yes 胞外 Extracellular OePRX106 EVM0051394 0 343 37.01 8.94 有 Yes 叶绿体 Chloroplast 2.2 Ⅲ类PRX基因的进化扩张
Ⅲ类PRX基因是一类植物特有的PRX基因,莱茵衣藻(Chlamydomonas reinhardtii)、小立碗藓(Physcomitrella patens)、江南卷柏(Selaginella moellendorffii)[31]、拟南芥[13]、毛果杨[16]、玉米[15]和水稻[14]中分别有6、31、62、73、93、119和138个Ⅲ类PRX基因,说明Ⅲ类PRX基因家族在进化过程中不断发生扩张,这可能与Ⅲ类PRX基因参与了植物广泛的生理过程有关。如图1,利用邻接法构建了包含73个AtPRX、93个PtPRX和106个OePRX蛋白的系统进化树,然后用同样的方法构建了仅包含106个OePRX蛋白的系统进化树,根据3种植物PRX蛋白的系统进化关系和106个OePRX蛋白之间的系统进化关系,将Ⅲ类PRX基因家族分为14个组。组1、2、4聚为1支,组6、7聚为1支,组9单独聚为1支,OePRX和PtPRX基因在这3支中均发生了明显的扩张;组3、10聚为1支,组12单独1支,OePRX和PtPRX基因在这2支中均发生了轻微的收缩。另外,OePRX基因在组13中发生了扩张,而PtPRX基因发生了收缩;PtPRX基因在组5、8、11中发生了扩张,OePRX基因在组5、11中发生了收缩。另外发现:组1 ~ 13中,OePRX、PtPRX、AtPRX基因均有分布;组14中仅包含10个OePRX基因,是一个油橄榄特有的组;这说明组1 ~ 13中,油橄榄、毛果杨、拟南芥有共同的Ⅲ类PRX基因的祖先基因,组14中的OePRX基因发生了明显的遗传分化。
2.3 OePRX基因的染色体定位和复制基因分析
2.3.1 OePRX基因在染色体上的不均匀分布
为了分析OePRX基因家族在进化过程中的扩张,绘制了106个OePRX基因在23条染色体上的分布图(图2)。结果发现:100个OePRX基因不均匀地定位于23条染色体上,6个OePRX基因定位于6个不同的contig上。第13条染色体(Chr13)上OePRX基因最多,数量为10(9.4%);其次是第6条染色体,数量为8(7.5%);第10条上有7个OePRX基因(6.6%),第11、12、15、23条上各有6个OePRX基因;第14、19条上各有5个OePRX基因;第3、7、9、17、20、22条上各有4个OePRX基因;第8条上有3个OePRX基因;第1、2、4、5、16、18、21条上各有2个OePRX基因。
2.3.2 OePRX共线性分析
油橄榄经历了2次木犀科(Oleaceae)特有的全基因组复制事件,分别发生在约28和59百万年前[1]。为了挖掘OePRX基因家族的进化扩张与油橄榄全基因组复制事件的关系,利用TBtools v1.095中的One Step MCScanX分析了OePRX基因家族内的复制基因,发现OePRX基因家族中共62个OePRX基因参与了基因复制,占OePRX基因总数量的58.5%。如图2所示:15个基因(14.2%)参与了串联复制,共得到了6个串联复制基因簇,6个基因簇分布于不同的染色体上,分别是第6、11、12、13、15、23条染色体。如图3所示:50个基因(47.1%)参与了片段复制,共得到了34对片段复制基因,34对基因分布于19条染色体上,第2、21、22条上未分布,说明片段复制是油橄榄Ⅲ类PRX进化扩张的主要驱动力。值得注意的是,3个基因(OePRX43、OePRX58、OePRX68)既参与了片段复制,也参与了串联复制。
2.3.3 拟南芥、毛果杨和油橄榄PRX基因同线性分析及Ka、Ks分析
为了进一步探究OePRX基因家族的进化扩张,绘制了OePRX与AtPRX和PtPRX基因的同线性图(图4)。结果表明:油橄榄和拟南芥之间有40对Ⅲ类PRX复制基因;油橄榄和毛果杨之间有76对Ⅲ类PRX复制基因。并注意到,25个OePRX(23.6%)基因在拟南芥和毛果杨中均有同源基因,推测在拟南芥、油橄榄和毛果杨分化前,双子叶植物可能有过一次基因复制事件,这与Tuskan等[32]的研究一致。另外,31个OePRX(29.2%)基因在拟南芥中有直系同源基因,46个OePRX(43.4%)基因在毛果杨中有直系同源基因,推测拟南芥与油橄榄的分化可能早于毛果杨与油橄榄的分化。
2.3.4 复制基因的Ka、Ks和Ka/Ks值分析
用KaKs_Calculator2.0计算复制基因的Ka、Ks和Ka/Ks值(图5)。在OePRX基因家族的6个串联复制基因簇和34对片段复制基因中,1个串联复制基因簇(OePRX97、OePRX98和OePRX99)和5对片段复制基因(OePRX8/OePRX66、OePRX16/OePRX26、OePRX24/OePRX61、OePRX36/OePRX96、OePRX35/OePRX100)的Ks在0.2 ~ 0.3之间,推测这些基因的复制可能发生于28百万年前的油橄榄全基因复制事件[1]。另外,发现油橄榄复制基因对、Oe-At复制基因对、Oe-Pt复制基因对的Ka/Ks均小于1,说明Ⅲ类PRX在双子叶植物的进化过程中,受到纯化选择的作用。油橄榄片段复制基因对的Ka/Ks小于串联复制基因对的Ka/Ks,说明纯化选择在片段复制基因中更强。
![]() 图 5 油橄榄Ⅲ类PRX各复制基因对的Ka、Ks分析Ka. 非同义替换;Ks. 同义替换;Oe-At. 油橄榄和拟南芥之间的PRX复制基因对;Oe-Pt. 油橄榄和毛果杨之间的PRX复制基因对。Ka, nonsynonymous substitution rate; Ks, synonymous substitution rate; Oe-At, PRX duplicate gene pairs between olive and Arabidopsis thaliana; Oe-Pt, PRX duplicate gene pairs between olive and black cottonwood.Figure 5. Ka, Ks analyses of olive class Ⅲ PRX duplicated genes
图 5 油橄榄Ⅲ类PRX各复制基因对的Ka、Ks分析Ka. 非同义替换;Ks. 同义替换;Oe-At. 油橄榄和拟南芥之间的PRX复制基因对;Oe-Pt. 油橄榄和毛果杨之间的PRX复制基因对。Ka, nonsynonymous substitution rate; Ks, synonymous substitution rate; Oe-At, PRX duplicate gene pairs between olive and Arabidopsis thaliana; Oe-Pt, PRX duplicate gene pairs between olive and black cottonwood.Figure 5. Ka, Ks analyses of olive class Ⅲ PRX duplicated genes2.4 OePRX基因序列分析
2.4.1 PRX基因基序分布和基因结构分析
如图6A,用邻接法构建了106个OePRX蛋白的系统进化树,OePRX蛋白家族分为14个组,组1、2、5、7、14中的OePRX蛋白明显多于其他组。106个OePRX蛋白平均约有324个氨基酸,使用MEME分析了每一个OePRX蛋白的基序分布(图6B),基序个数设置为10,每个基序为6 ~ 50个氨基酸。发现46个OePRX蛋白(43.4%)含有10个基序,34个OePRX蛋白(32.1%)含有9个基序,剩下的OePRX蛋白含有3 ~ 8个基序,说明OePRX蛋白的基序分布整体比较保守。另外还注意到,组1 ~ 13中OePRX蛋白含有6 ~ 10个基序PRX,组14的OePRX蛋白含有3 ~ 5个基序,进一步说明组14中的OePRX蛋白可能发生了功能分化。
然后,用GSDS绘制了OePRX基因的基因结构图(图6C),OePRX基因中有0 ~ 12个内含子不等,其中,84个OePRX基因(79.2%)中有2 ~ 4个内含子。PRXOePRX基因的大多数内含子位置保守,为0,且所有OePRX基因第1个内含子的位置最为保守,均为0。此外,同组中OePRX基因内含子的数量和位置相对保守,需特别注意的是,组14中OePRX基因中的内含子数量(6 ~ 12个不等)明显多于其他组(0 ~ 4个不等,除OePRX32、OePRX43、OePRX67外),且内含子位置多变,暗示组14中Ⅲ类PRX基因可能具有特殊的进化模式和不同于其他组OePRX基因的功能。
2.4.2 OePRX启动子上顺式作用元件分析
利用OePRX基因转录起始位点上游1 500 bp作为启动子预测顺式作用元件,共预测到59个顺式作用元件(图7)。CAAT-box控制转录起始的频率,TATA-box是RNA聚合酶的结合位点之一,这2个顺式作用元件在106个OePRX基因的启动子中均有分布。34个顺式作用元件(57.6%)与发育有关,其中,光应答顺式作用元件Box 4、G-box、GT1-motif和TCT-motif广泛分布于OePRX基因的启动子中,这说明OePRX基因在油橄榄生长发育过程中具有重要作用。17个顺式作用元件(28.8%)与激素和胁迫应答有关,包括脱落酸(ABA)、水杨酸、赤霉素、茉莉酸甲酯(MeJA)、生长素、低温、干旱、缺氧等,ABRE是依赖ABA表达基因的顺式调节元件,分布于81个OePRX基因的启动子中;CGTCA-motif和TGACG-motif是依赖MeJA表达基因的顺式作用元件,在64个OePRX基因的启动子中有分布;ARE、GC-motif均为厌氧胁迫应答的顺式作用元件,其中ARE分布于76个OePRX基因的启动子中,说明OePRX基因广泛参与激素代谢过程,调控油橄榄的生长发育,且能响应逆境胁迫,增强油橄榄抗逆性。
2.5 OePRX表达模式分析
2.5.1 不同器官或组织中OePRX的差异表达
为了研究OePRX基因家族的表达模式,分析了106个OePRX在根、茎、叶、花、果和分生组织中的转录组数据,发现在106个OePRX基因中,61个OePRX基因有表达,tpm在1 ~ 846之间;其余OePRX基因的tpm小于1,可认为其不表达。如图8所示:根据表达模式的相似性,106个OePRX基因在不同组织中的表达量聚为8个模块(module)。18个基因(17.0%,模块A、B和C)在不同组织内普遍表达,其中7个基因(模块A)的整体表达量较高,5个基因(模块B)表达量次之,6个基因(模块C)表达量较低。13个基因(12.3%,模块D、E、F和G)在1个或多个特定的组织内表达,其中,5个基因(模块E和F)在除果实之外的其他组织内表达,2个基因(模块E)在茎、叶、分生组织中表达量更高,7个基因(模块D)主要在花中表达,1个基因OePRX72(模块G)主要在根中表达。值得注意的是,75个基因(70.8%,模块H)几乎不表达或者仅在特定的组织中具有极低的表达。
![]() 图 8 油橄榄Ⅲ类PRX基因在不同器官或组织中的表达模式模块A. 在不同组织中均高表达;模块B. 在不同组织中均有较高表达;模块C. 在不同组织中均较低表达;模块D. 在花中特异高表达;模块E. 在除果外的组织中高表达;模块F. 在除果外的组织较高表达. 模块G. 在根中特异高表达;模块H. 在各组织中低表达或几乎不表达。Module A, high expression in all tissues; module B, relative high expression in all tissues; module C, relative low expression in all tissues; module D, high specific expression in flower; module E, high expression in all tissues except fruit; module F, relative high expression in all tissues except fruit; module G, high specific expression in root; module H, low or no expression in all tissues.Figure 8. Expression patterns of olive class Ⅲ PRX genes in different organs or tissues
图 8 油橄榄Ⅲ类PRX基因在不同器官或组织中的表达模式模块A. 在不同组织中均高表达;模块B. 在不同组织中均有较高表达;模块C. 在不同组织中均较低表达;模块D. 在花中特异高表达;模块E. 在除果外的组织中高表达;模块F. 在除果外的组织较高表达. 模块G. 在根中特异高表达;模块H. 在各组织中低表达或几乎不表达。Module A, high expression in all tissues; module B, relative high expression in all tissues; module C, relative low expression in all tissues; module D, high specific expression in flower; module E, high expression in all tissues except fruit; module F, relative high expression in all tissues except fruit; module G, high specific expression in root; module H, low or no expression in all tissues.Figure 8. Expression patterns of olive class Ⅲ PRX genes in different organs or tissues对比图8中OePRX基因的进化分组和表达模块发现:同一组中的基因表达模式相似,比如组3、7、8、9、10和12中的基因均有单一的基因表达模块;也有一些组中的基因表达模式的变化幅度比较小,比如组2、4、6和11中的基因均聚类为2个表达模块。然而,组5中的基因聚类为6个不同的表达模块,组1、13、14中的基因均聚类为3个不同的表达模块。尽管如此,在组织间普遍表达类模块A、B和C的基因其进化关系更为相近,在特定组织表达类模块D、E、F和G的基因也是如此。另外,值得注意的是,模块H中几乎不表达或者仅在特定的组织表达的75个基因均匀分布于各个组。
2.5.2 实时荧光定量PCR验证
运用实时荧光定量PCR验证了6个基因(OePRX1、OePRX8、OePRX33、OePRX36、OePRX66和OePRX99)在叶片、茎、根和果实中的表达量(图9)。运用Primer Premier5.0设计引物,如表2所示。RNA-seq数据和实时荧光定量结果均表明,OePRX1和OePRX36基因在根中表达量最高,在果实中表达量最低;OePRX8和OePRX66基因在叶片和果实中表达量较高,在茎和根中表达量较低;OePRX33基因在茎和根中表达量较高,在叶和果中表达量较低;OePRX99基因在茎中表达量最高,在果实中表达量最低。
表 2 6个油橄榄Ⅲ类PRX基因RT-qPCR的引物Table 2. Primers of six olive class Ⅲ PRX genes used for RT-qPCR基因名称
Gene name引物(5′—3′) Primer sequence (5′−3′) 基因产物长度
Gene product length/bpOePRX1 F: TCCAGGAGTTGTTTCTTGTGC 114 R: CCTTCTTCCGTCTTTTCTTCC OePRX8 F: AAGGGCGCATCCTGAAAG 200 R: AGAAGGCATCTTCATCCTTAGC OePRX33 F: GGAAGCATAATAAGCGAGAAG 235 R: TTATTGGAGCCACTCAAACTG OePRX36 F: ACAAACGCCACAAGAACACTG 195 R: CACTCCCTCTCTAAAGCCTCC OePRX66 F: GATTCGCAATGAGGAGGAGTA 140 R: AGCAACAACACCAGCAAGC OePRX99 F: GCTCGGGCTTTTAGAATCATC 204 R: GAAGGAAGGTTTGCCAGTGTT 2.6 非生物胁迫下OePRX基因的差异表达
为了研究油橄榄Ⅲ类PRX基因对非生物胁迫的响应,利用RNA-seq数据分析了油橄榄响应干热胁迫和水涝胁迫过程中差异表达的OePRX基因。如图10所示:干热胁迫下,39个OePRX差异表达,占比36.8%;水涝胁迫下,10个OePRX差异表达,占比9.4%。基于差异表达基因在干热胁迫和复水过程中的表达趋势,将其分为4个模块:模块1,23个基因(21.7%)的表达在干旱胁迫过程中显著下调,复水后显著上调;模块2,7个基因(6.6%)的表达在干旱胁迫过程中显著上调,复水后显著下调;模块3,3个基因(2.8%)的表达在高温和前期干旱时变化不显著,在后期干旱时显著上调,复水后显著下调;模块4,6个基因(5.7%)的表达在高温胁迫下显著上调,解除高温胁迫后,显著下调。另外,与正常浇水的油橄榄相比,在第13天,高温和早期干旱处理的油橄榄中有8个基因(7.5%)显著上调,分别是:OePRX31、OePRX36、OePRX45、OePRX46、OePRX63、OePRX82、OePRX83和OePRX96。在第27天,13个基因(12.3%)显著上调,分别是:OePRX2、OePRX16、OePRX26、OePRX31、OePRX33、OePRX36、OePRX63、OePRX76、OePRX79、OePRX82、OePRX96和OePRX104;4个基因(3.8%)显著下调,分别是OePRX40、OePRX50、OePRX65和OePRX99;在第80天,OePRX基因表达无显著变化。水涝胁迫处理90 d后,油橄榄叶片中10个OePRX基因(9.4%)的表达显著下调,猜测这些差异表达的Ⅲ类PRX在油橄榄响应非生物胁迫胁中具有十分重要的作用。PRX除干热和水淹胁迫外,我国油橄榄常常面临低温和营养等胁迫,因此有必要对其进行更为丰富而广泛地探索,进一步研究Ⅲ类PRX基因在油橄榄响应非生物胁迫过程中的功能。
![]() 图 10 油橄榄Ⅲ类PRX基因响应干热和水涝胁迫的差异表达模式Day(−3). 干旱处理前3天;Day13. 干旱处理第13天且伴随高温;Day27. 干旱处理第27天;Day80. 复水后52天。模块1. 干旱期间基因表达下调,复水后上调;模块2. 干旱期间基因表达上调,复水后下调;模块3. 后期干旱期间基因表达上调,后期干旱及复水后下调;模块4. 仅在高温下基因表达上调,高温胁迫后,基因表达下调。复制基因名称的颜色与系统进化分组的颜色相对应。Day(−3), 3 d before drought treatment; Day13, 13th day of drought treatment with high temperature; Day27, 27th day after drought treatment; Day80, 52 d after re-irrigation. Module 1, gene expression is down regulated during drought and up regulated after rehydration; module 2, gene expression is upregulated during drought and downregulated after rehydration; module 3, gene expression is upregulated during late drought, and downregulated after late drought and rehydration; module 4, gene expression is upregulated only under high temperature, and downregulated after high temperature stress. The color of the replicated gene name corresponds to the color of the phylogenetic grouping.Figure 10. Differential expression patterns of class Ⅲ PRX genes in olive oil in response to dry heat and waterlogging stresses
图 10 油橄榄Ⅲ类PRX基因响应干热和水涝胁迫的差异表达模式Day(−3). 干旱处理前3天;Day13. 干旱处理第13天且伴随高温;Day27. 干旱处理第27天;Day80. 复水后52天。模块1. 干旱期间基因表达下调,复水后上调;模块2. 干旱期间基因表达上调,复水后下调;模块3. 后期干旱期间基因表达上调,后期干旱及复水后下调;模块4. 仅在高温下基因表达上调,高温胁迫后,基因表达下调。复制基因名称的颜色与系统进化分组的颜色相对应。Day(−3), 3 d before drought treatment; Day13, 13th day of drought treatment with high temperature; Day27, 27th day after drought treatment; Day80, 52 d after re-irrigation. Module 1, gene expression is down regulated during drought and up regulated after rehydration; module 2, gene expression is upregulated during drought and downregulated after rehydration; module 3, gene expression is upregulated during late drought, and downregulated after late drought and rehydration; module 4, gene expression is upregulated only under high temperature, and downregulated after high temperature stress. The color of the replicated gene name corresponds to the color of the phylogenetic grouping.Figure 10. Differential expression patterns of class Ⅲ PRX genes in olive oil in response to dry heat and waterlogging stresses3. 讨论与结论
Ⅲ类PRX基因通过氧化特定小分子、调节活性氧水平来促进木质化、细胞伸长、响应生物胁迫和非生物胁迫[33-34]。为了调控基因表达的时空特异性、蛋白分布的特定性和过氧化物酶活性的多样性,高等植物进化得到了庞大的Ⅲ类PRX基因家族[35]。在高等植物的进化过程中,Ⅲ类PRX基因家族通过全基因组复制事件、片段复制、串联复制和转座子复制等方式不断扩张。前人研究表明:拟南芥中有73个PRX基因,9对片段复制基因,10个串联复制基因簇[13];毛果杨中有93个PRX基因,由60 ~ 65百万年前的全基因组复制事件产生了14对复制基因,共有37个串联复制基因簇[16];白梨中有94个PRX基因,26对片段复制基因,1个串联复制基因簇[17];水稻中有138个PRX基因,多于20对的片段复制基因和6个串联复制基因簇[14];玉米中有119个PRX基因,16对片段复制基因和12个串联复制基因簇[15]。本研究中,在双子叶木本植物油橄榄中鉴定得到了106个Ⅲ类PRX基因,其中,62个OePRX(58.5%)参与了基因复制,共产生了6个串联复制基因簇和34对片段复制基因。据此猜想,片段复制可能是高等植物PRX基因家族进化扩张的主要方式。
基因复制事件是基因组进化过程中基因功能冗余和分化的主要来源[36]。复制基因的功能具有相关性,且其功能的分化与其表达模式的分化有关。例如:三倍体小麦中的同源基因VRN-A1、VRN-B1和VRN-D1是调节开花时间的重要因子,不同抽穗期的小麦中,这3个同源基因在不同组织中的表达量不同[37]。本研究发现,许多OePRX基因在不同组织中的表达模式与其在拟南芥或其他模式物种中的表达模式相似,说明PRX的基因功能在不同植物之间具有保守性。AtPRX33和AtPRX34基因是一对高度同源(95%)的Ⅲ类PRX基因,但是其启动子和内含子序列却高度分化。半定量RT-PCR结果表明,这2个基因主要在拟南芥幼苗的根部表达,并且光照对其表达具有促进作用。与野生型拟南芥相比,AtPRX33基因突变体的根变短,AtPRX33和AtPRX34基因双突变体的根更短,而AtPRX34基因过表达拟南芥幼苗的根显著变长[38]。本研究发现:OePRX基因家族中的6个串联复制基因簇和34个片段复制基因的分子量、等电点、信号肽的分布和长度以及基序分布均比较保守;但2个串联复制基因簇(33.3%)和22对片段复制基因(64.7%)的内含子位置发生分化;1个串联复制基因簇(16.7%)和11对片段复制基因(32.4%)在不同组织中的表达模式发生分化。因此,有必要利用分子生物学手段深入研究油橄榄Ⅲ类PRX基因家族内复制基因的亚功能和新功能。
为了研究单个Ⅲ类PRX基因在逆境下的特定功能,大量Ⅲ类PRX转基因植株已经获得。过表达甜土豆(Ipomoea batatas)swpa4 PRX基因的烟草(Nicotiana tabacum)幼苗和过表达AtPRX3基因的拟南芥幼苗[39]的耐盐性和耐旱性增强。过表达AtPRX22、AtPRX39和AtPRXrx69基因的拟南芥对油菜素类固醇不敏感,而突变体的耐寒性增强[40]。同样地,2个长春花(Catharanthus roseus)PRX基因在烟草中的异源转化提高了其在脱水胁迫下的发芽率,同时提高了其耐旱性[41]。辣椒(Capsicum annuum)CpPRX2的转基因拟南芥幼苗提高了其体内H2O2水平的积累,同时提高了其耐病性、耐旱性和耐盐性[42]。菜豆(Phaseolus vulgaris)过氧化物酶FBP1反义表达的拟南芥中,AtPRX33和AtPRX34基因的表达量降低,体内氧化应激受到抑制,抗病性减弱[43]。本研究发现:干热胁迫和水涝胁迫下油橄榄中40个OePRX基因(37.7%)显著差异表达,且差异表达模式多样(图10),猜测其他OePRX基因可能参与另外的生物胁迫或非生物胁迫,需要对多种胁迫下的油橄榄幼苗开展更为广泛的研究,锁定响应胁迫的关键基因,从而深入挖掘其功能。Wu等[44]研究表明:经历了多次多倍化复制的植物保留了许多胁迫相关的基因家族,且得到的复制基因通常是胁迫相关通路的关键因子。本研究结果表明:与正常生长的油橄榄相比,干热胁迫下油橄榄中串联复制基因簇(OePRX58和OePRX59)和3对片段复制基因(OePRX16和OePRX26,OePRX36和OePRX96,OePRX37和OePRX79)的表达量均显著降低,片段复制基因对(OePRX22和OePRX64)的表达量均显著上升,片段复制基因对(OePRX1和OePRX45)的表达量仅在高温下显著上升;水涝胁迫下,片段复制基因对(OePRX36和OePRX96)的表达量均显著降低,猜测这些复制基因之间具有较强的功能相关性,可能是研究油橄榄非生物胁迫响应通路的关键候选基因。
综上所述,油橄榄OePRX基因家族明显的进化扩张和多变的表达模式暗示了其功能的复杂性,尤其是复制基因的亚功能化和新功能化对油橄榄在地中海地区的广泛分布具有十分重要的适应性意义。
-
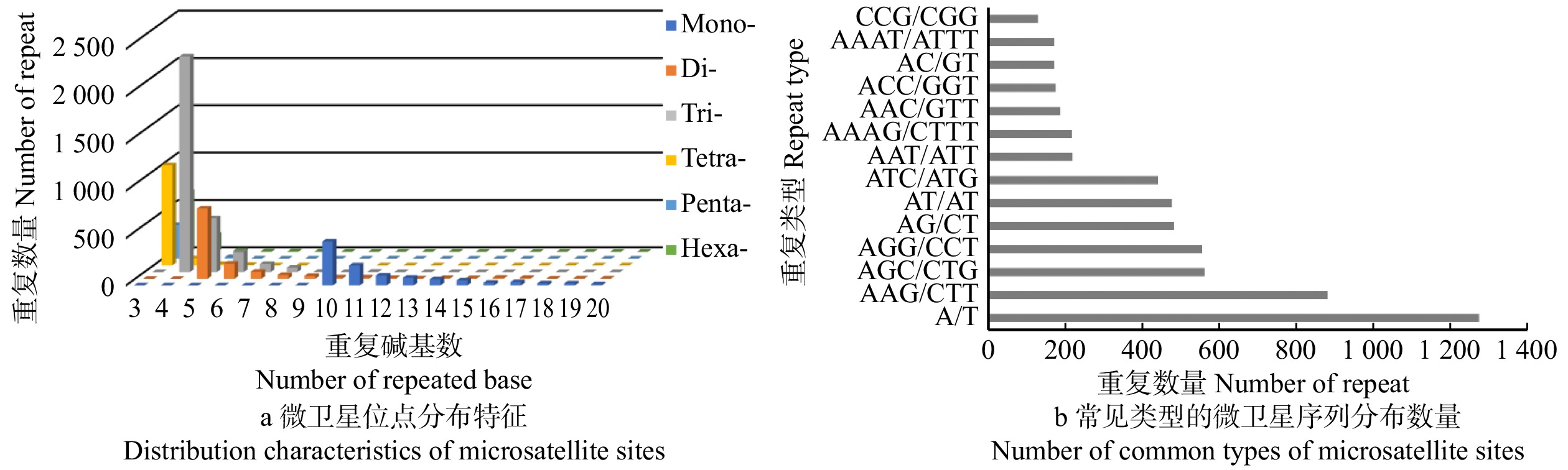
图 1 白皮松不同类型的EST-SSR重复序列的分布频率
Mono-. 单碱基重复基元;Di-. 二碱基重复基元;Tri-. 三碱基重复基元;Tetra-. 四碱基重复基元;Penta-. 五碱基重复基元;Hexa-. 六碱基重复基元。Mono-, one-base-repeat motif; Di-, two-base-repeat motif; Tri-, three-base-repeat motif; Tetra-, four-base-repeat motif; Penta-, five-base-repeat motif; Hexa-, six-base-repeat motif.
Figure 1. Distribution frequency of EST-SSR repeats in different types of P. bungeana
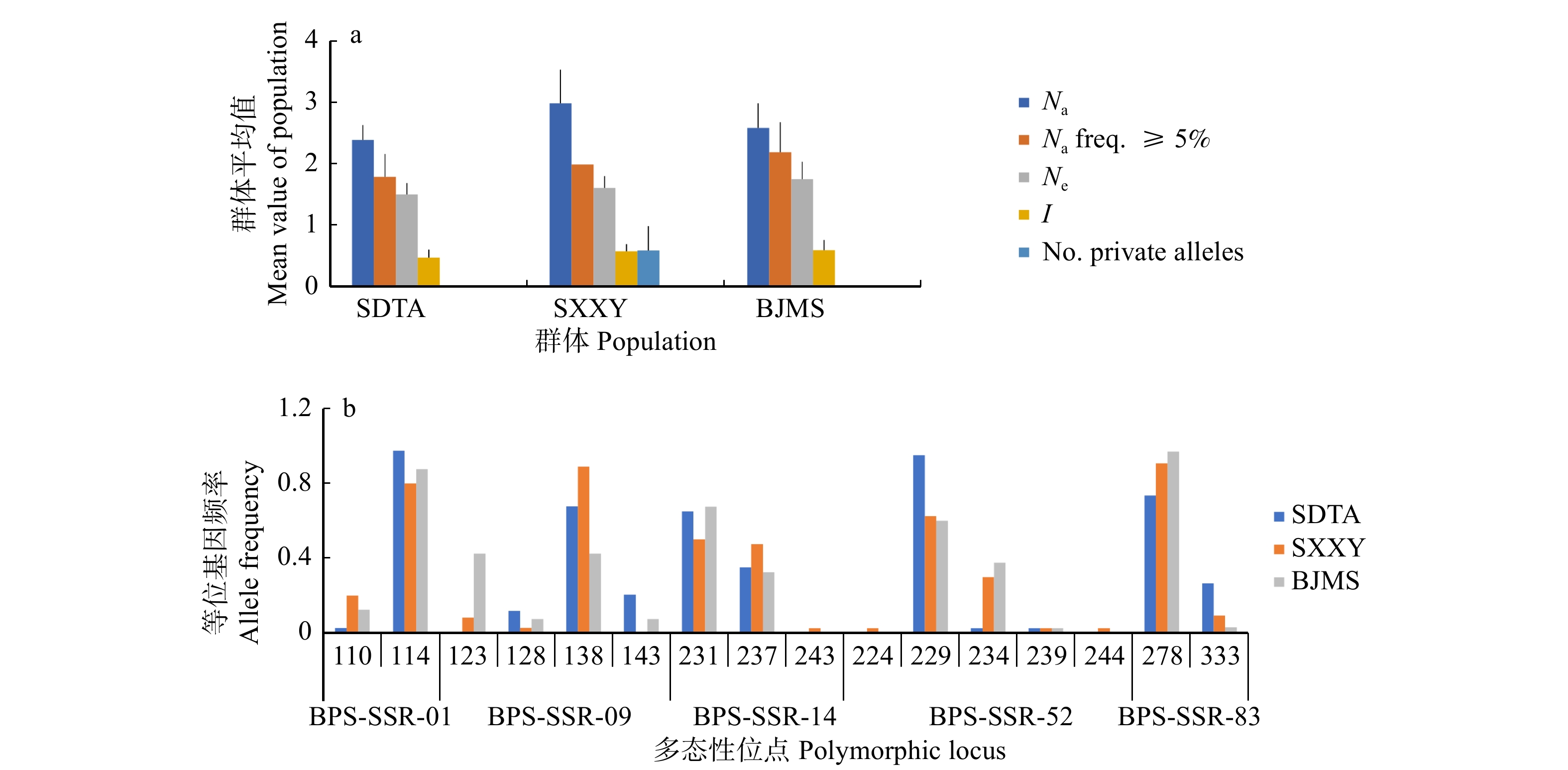
图 3 5个微卫星位点在3个群体中的等位基因分布模式
Na. 等位基因数量;Na freq. ≥5%. 基因分布频率大于等于5%的等位基因数量;Ne. 有效等位基因数量;Ⅰ. 香浓多样性指数;No. private alleles. 单个群体中特有的等位基因数量。Na, number of alleles; Na freq. ≥5%, number of alleles whose gene distribution frequency is greater than or equal to 5%; Ne, number of effective alleles; Ⅰ, Shannon’s diversity index; No. private alleles, number of alleles unique to a single population.
Figure 3. Allelic distribution patterns of five microsatellite loci in the three populations
表 1 白皮松3个群体的地理位置及主要生态因子
Table 1 Geographical location and main ecological factors of three populations in Pinus bungeana
林分代号
Abbreviation of
stand样本数量
Sample size林分来源
Stand source经度
Longitude纬度
Latitude海拔
Altitude/m年降水量
Annual
precipitation/mm林分起源
Stand originSDTA 20 山东泰安 Taian, Shandong 116°17′E 36°07′N 61 600.0 天然林 Natural forest SXXY 20 山西孝义 Xiaoyi, Shanxi 111°48′E 37°12′N 950 493.0 天然林 Natural forest BJMS 20 北京蟒山 Mangshan, Beijing 116°16′E 40°15′N 330 631.0 人工林 Plantation 表 2 白皮松EST-SSR引物
Table 2 Primers used for EST-SSR analysis of P. bungeana
引物编号 Primer No. 前引物序列 Forward primer sequence 后引物序列 Reverse primer sequence 元件 Motif BPS-SSR-01 CATAACACAGCAGCAACCGG ACACTGCTGCAAAAATAACCCT A BPS-SSR-02 GCCGAAACGACCTTTCACAC CTAAAGCATAAACCGCCGCC AAAT BPS-SSR-03 TCCAAGAAAGCGCGTTCTCT TGTTATTTTATTAGCGCGGCCG GAG BPS-SSR-04 ACAAGGGGAGAGCAATGTGG GCCATTTCCAACAGCTTAGCA GA BPS-SSR-05 TGAGAAGAAGGCCGAGGAGA GCTTGAGCACCTTGAAGATGT AAG BPS-SSR-06 AGCCCTAGTCCGAGTCCAAT GAGGCCCAGGAGGAGAAAAC CTG BPS-SSR-07 TGCCAGAGACAATGCAGACA GGAAATACCTGTCGGCCACA TGA BPS-SSR-08 TCTGGTGTTGTGGATGCCTC ACGCTGCTGTTACTTCCGAA GAAAG BPS-SSR-09 CGGCCTCAACAACCTCTTCT TCCTCCGGCTGTTCGTAGTA CTCTC BPS-SSR-10 AGCCGAAATCAAAAGCGCAG GGGGACAACGCGGATGAA CCTGAC BPS-SSR-11 GGCGTGGTGGTGAATTGAAC GAGCTTGGGGGCAATCTTCT CTC BPS-SSR-12 TTCACCCCCTGCAGATTTCC GCAAGCCTCCCCTTCAGAAT ATTCA BPS-SSR-13 AAGGGCAAGAGTAAACGGGG GTTGGCGGCATTGCAGAATC GAGGAA BPS-SSR-14 CTGGGTGTTCCTACCAGCAG ACCCTCAGTTTGTCAGGGGA TGATAA BPS-SSR-15 TACAGTAGGAAGCAGTGCCA GAGGGAGTGGCGTTATCGAG GGCAAC BPS-SSR-16 CTGCAGATGAGGAGGCTGAC AGCAGCAACCTAGAGGCAAT GAA BPS-SSR-17 ACAGAAAGTGGGAGGAAGCG TCTCCTCTCCCCTCCTCTCA GAAGA BPS-SSR-18 CTCCTGGCCATCCCAAACAA AACTACGAAAGCTCCAGGCC GGTGAT BPS-SSR-19 CAGCTCAACTCGCATTACGC ACCACCTGTGTTTGGCAAGA GCA BPS-SSR-20 TGGGATTGTCCAAGAGCAGG TCACTCTGATTGTGACTGCGA CAATCC BPS-SSR-21 GGATGCAGAGGACGAGAGG TCGGATCTCCTCTTCCCCTG AGGGGA BPS-SSR-22 TGCTCCACAGACAGAAGCTG CCCCTAGCATTGTTGTACCTTC GAT BPS-SSR-23 CATTCTTGGGGGCTCTACCG TAAACGGCCTGACTCCAACC AT BPS-SSR-24 TCCAGGTGGCTTTGGTTCTC GCAGGTCACTGGTATCGACC T BPS-SSR-25 CACAGGGAGTGGCGGTATAC AGCATCGATTCCTCCACTGC GGCGAT BPS-SSR-26 GGTGACCCTTTGGAGAGGTC AAAATCGCCACCATTGCCAC CCACA BPS-SSR-27 TTGTGTGAGCAGCCAACAGA GGGGAGATCAATGACAACCCA TTCCTC BPS-SSR-28 TACCTCCATGGGGGTGTTGA GCTCTGGACTGGATGGTCAG CAGAG BPS-SSR-29 ATTGGGAGGGCGGAGAAAAG GGCGGCATTGACAATGAGAC GGCAGA BPS-SSR-30 GCTTATTCCCTGGGCATGGT CTGTGGCCAAGCATTGCTTC CAGCGA BPS-SSR-31 TCTCTCACGCGTTTTGCAGA AGAAGTGGTCCACTCCCGTA TGATG BPS-SSR-32 GCGATTGATCAGGATGCACC TGCTGAATTTTTGCAGCAGAGA AT BPS-SSR-33 TCGCTCGTACTTGCACCATT TCGGACTCAGGCCAGTACTT TCTTC BPS-SSR-34 TCAAAACGGCACTGGCATTG CCGATCAATGGCCATGGCTA TGGCTT BPS-SSR-35 TCCCTGCACAGCTGAAAACA AATGACAATGCTTTGGCCGG ACTGCG BPS-SSR-36 CTCTGTTCCGTCGGTGATCC GCTCTTCCAACTTGCGCTTC GGACAT BPS-SSR-37 TTTGGAGCAATGTTTCGGCG ACACTGCATCCCACTGTCTG ATAGAG BPS-SSR-38 ACTTCGTGAGCAGGTTGAGG AATTCCTCTGCTTCGGGTGC AAG BPS-SSR-39 TTACTGACAGGTCGCCTTCG TATATGGGTGGCTGCCTCTG ATGTCG BPS-SSR-40 ACCACGTCATCATCGTCGTC AAACTGAGGCAGAGGCTTCC GAGGCA BPS-SSR-41 ATTATGGGCGGTGGCAGAAA CCCCGTTCCTACCATCCAAT GAA BPS-SSR-42 GGAATCTCAGCGCTTCCTGT AAGCTCTCACAAACGTGGCA CAG BPS-SSR-43 CTGTCGAGCGAAGGTTACCC AGGTCCCTTGCCCATCTCTA GCAGTT BPS-SSR-44 GGGTGAGCTAGACCGTCAAA TATTGACGGCGGCCTTTCTT CTCTG BPS-SSR-45 CCGCGAAGACAGAGTATCCC AGCTGGGAAAAAGATCGCCA TAT BPS-SSR-46 GAAAATTGCCCCCGTCGAAG GCAAAGACGCTTCCAGCAAA GAGGAA BPS-SSR-47 GCCTTATCTTCTTCCTTGTGTGA CACATAGAAGAGGCGGCAGT TCT BPS-SSR-48 GGAAGGTTGCCAGGTGGTAA ACATCACAACGAAGGGACCC TGGAGA BPS-SSR-49 CTCGAGTCTCTCACCCTTGC CCGTGTTTGTCCACCAGACT AGCCGC BPS-SSR-50 ACGCCGGATTTCAACAGACT GGGGAGAACGGCGATTGTTA AACGCC BPS-SSR-51 AAGGGCTCCATGATGTCTGC ACGTGGGCATAGGCTTGAAA CTTCAC BPS-SSR-52 ATCTTTGTCCAGCGCTTCGA GGAAGCACGGGAATCAGACA CCTCT BPS-SSR-53 ATCAGTACAACGCTGCCCAG CTGCCCGGAGTCTGAGAATC CAG BPS-SSR-54 GTCTTTCGGTGTGGGCTGTA CCGTCACACGATCGTCCAAT TTGT BPS-SSR-55 TGCAGGGAGTGTCTACAGGA GAGAAACGTGAAGCCCCTCA GAC BPS-SSR-56 CCTTTGCTCTGTACGAGGGG TCAGTCGTTTCAGTCGCAGT GCACGG BPS-SSR-57 TGGGGGAAAAGCTAGATCCA CTTTGTGTTTGGCCTCCAGC AGAGGC BPS-SSR-58 TGAGGGCAATGTTACTCGCT AACAATCGCCGCTATCAGCT AGG BPS-SSR-59 AGGGGCCTCAGCTGATATCA AACAACAGCAACAGCAGCAG TGC BPS-SSR-60 CTATCAGGGGATTGTGGGGC TCTGCTGCTGGAGACGTAAC CGG BPS-SSR-61 TATTCGCCACTCTGCACGTT CCTCCATGGGGAAGGGAATG TTGCAG BPS-SSR-62 ATTGATCAGGCGAGGCAGAC GCAGGTTTTCGTTGTGGGTG GGGCGA BPS-SSR-63 ATCGAACCCGTAGTCTCCCA AAGTCAAGAAAGTGGCCGCT TGGGCT BPS-SSR-64 CCGCGGTTTTGATGTGAACC CCCTGTCCAGTTCGTTGTCA GACAAC BPS-SSR-65 GGCTAGCGAGTGGTGATTGT GAGTTGCTTCTCCTCCGCAT CCTGGC BPS-SSR-66 TTGCGAAGGAGGTCCATAGC GGGGTAGATGGAGAATGGCC AAAGGC BPS-SSR-67 GCTTCGTTGGTAAGGGCCTA AGTTCTTGCACTGCCCAACT GATGGA BPS-SSR-68 GCCTTGTAGTTTTGGCCTGC CTGCAGGATGGGGCTCTTTT AGA BPS-SSR-69 AAGTCCTCGGCTTCGCATTC GAGCCCAGTGCCTCAATCTT CTT BPS-SSR-70 CGGGCAACAGTCTGTTAGGT GCCTGAAACAGCCTCCTCAT ATGGGC BPS-SSR-71 TAAGAAGCCTGCGTGGACTG GCATATTTGAATGCCTTTCTGGG AT BPS-SSR-72 CTCACCCGACAACTACAACA TGAGGGCTCTTGTTGACAGT CCCCTG BPS-SSR-73 CGTACCAAGTGCGCCTCTAT TAGTGCCAACTCTGCGCTAC GAG BPS-SSR-74 ACTGGGGGCAACAATATCAGA GAGTGGTGGGTGTGGAAGAG AGTGA BPS-SSR-75 CCTGCAACTGGATGACGACT ACCGGCACACAGAAAAGCTA CCACGA BPS-SSR-76 GCAGTCCCCTCGCTATAACC TCAGAAACCCAGCGGTGTAC AAATAA BPS-SSR-77 GAAGTAGAAGCAGGCGGAGG ATAACGCTGCCAGTGCTTCT AGCAGA BPS-SSR-78 GCTGATCCTCTTGACTCCGG GGCAATGGTGAGAGTGTGGA TGACCC BPS-SSR-79 GCCTAGGCACTGGAATTCCT GGTGGGCATGACAGATGGAA TTCCTC BPS-SSR-80 TGAGCTTTCACAACACCCGA GGCCCAATTCTCCTCCTTCC TGAT BPS-SSR-81 CCTCAGCTCATCCTACAGCG TCTCTTCCAGTTCGACTTCCA AAGGTG BPS-SSR-82 ACGGGTCATTGCGCCATTAT CCACGCCAACATTGACCTTG ATGGCG BPS-SSR-83 TTGTTCAATCCCTCCACCGC GGAATCTTCCCGTCCTGCAA TTGTT BPS-SSR-84 GGCCCTATGAGTGCACTGTT ACAGCCCCTACTATTGCGAA CAGGG BPS-SSR-85 GGGGATTCTTCAATCTGTAACGC ACCCCACGTAACCCTTATGC GATCCT BPS-SSR-86 TAGATACTCCGGTGCCAACG ATTTCAGCAGCCCATCCGAG CAG BPS-SSR-87 AAGAAAATGGTGGGCCGGAT CTTCCTCCAGCTCCCCAAAG AGCAGT BPS-SSR-88 TCTACACGGTCCAAGCGATG TTCCCTTTCGATGACCTGCA GAGCAG BPS-SSR-89 TGTCGAGAAGGCTATTGCTGA TGACATCTGCAACGTCTCCT ATA BPS-SSR-90 GGAAGCTTTGCATGTGCCAA GCAACCAGCTCTTCCGACTT TTTCA BPS-SSR-91 ATGGACATGCGTTGTGGAGT CTGGATACACATGGCCTCCC AGAGTG BPS-SSR-92 CGAACGTCTCCGGATCGAAT AGGGGAAGGCATGATTGTGG TCCTCT BPS-SSR-93 TCCGCCTGAGTTCCAAGTTC ACGGTGATGAAGGGAAAACCA A BPS-SSR-94 GATCTTCAGAGAGCTCCGGC ACCACTGGAAACACTTCGCA AAT BPS-SSR-95 GAGCATCGTCCGAATGTGGA TTGCCCCTCCTCTTCTTCCT AGA BPS-SSR-96 AAGCTGCTGAATTGGGCTCT ACACATGATGACAGGGCAGG TCTTGT 表 3 PCR反应体系
Table 3 PCR reaction system
μL 类别
Category预试验反应体系
Pre-experiment
reaction systemSSR-PCR反应体系
SSR-PCR reaction
system2 × PCR MIX 10.0 10.0 DNA模板 DNA template 2.0 2.0 前引物 Forward prime 3.2 0.8 后引物 Reverse prime 3.2 3.2 M13引物 M13 primer 3.2 ddH2O 1.6 0.8 总计 Total 20.0 20.0 表 4 白皮松中25个微卫星位点的特征
Table 4 Characteristics of 25 microsatellite loci developed in P. bungeana
引物编号
Primer No.等位基因数
Number of alleles (Na)元件
Motif观察杂合度
Observed
heterozygosity (Ho)期望杂合度
Expected
heterozygosity (He)多态性信息含量
Polymorphism information
content (PIC)BPS-SSR-01 2 A 0.203 0.211 0.187 BPS-SSR-09 4 CTCTC 0.364 0.530 0.484 BPS-SSR-14 3 TGATAA 0.433 0.487 0.374 BPS-SSR-52 5 CCTCT 0.417 0.423 0.361 BPS-SSR-83 2 TTGTT 0.265 0.232 0.204 BPS-SSR-62 2 GGGCGA 0.033 0.033 0.032 BPS-SSR-96 2 TCTTGT 0.000 0.130 0.120 BPS-SSR-04 1 GA BPS-SSR-08 1 GAAAG BPS-SSR-12 1 ATTCA BPS-SSR-16 1 GAA BPS-SSR-30 1 CAGCGA BPS-SSR-50 1 AACGCC BPS-SSR-54 1 TTGT BPS-SSR-63 1 TGGGCT BPS-SSR-64 1 GACAAC BPS-SSR-75 1 CCACGA BPS-SSR-79 1 TTCCTC BPS-SSR-82 1 ATGGCG BPS-SSR-90 1 TTTCA BPS-SSR-91 1 AGAGTG BPS-SSR-93 1 A BPS-SSR-95 1 AGA BPS-SSR-80 2 TGAT BPS-SSR-13 2 GAGGAA 表 5 3个白皮松群体遗传多样性参数
Table 5 Genetic diversity parameters of the three populations in P. bungeana
群体
PopulationNa 有效等位基因数量
Number of effective allele (Ne)香农多样性指数
Shannon’s diversity index (I)Ho 固定指数
Fixation index (Fix)SDTA 2.400 1.516 0.484 0.295 −0.026 SXXY 3.000 1.620 0.589 0.362 −0.075 BJMS 2.600 1.761 0.606 0.343 0.081 均值 Mean 2.667 1.632 0.560 0.333 −0.007 表 6 白皮松中不同SSR位点的F-统计量
Table 6 F-statistic of SSR locus in P. bungeana
位点 Locus 亚群内 Within subgroup (Fis) 群体内 Within group (Fit) 亚群间 Between subgroups (Fst) 基因流 Gene flow (Nm) BPS-SSR-01 −0.021 4 0.028 6 0.048 9 4.859 4 BPS-SSR-09 0.189 6 0.312 4 0.151 6 1.399 6 BPS-SSR-14 0.082 9 0.102 7 0.021 6 11.340 0 BPS-SSR-52 −0.123 6 0.006 0 0.115 3 1.918 1 BPS-SSR-83 −0.257 6 −0.149 3 0.086 1 2.652 0 均值 Mean 0.011 4 0.100 6 0.090 2 2.521 2 表 7 群体间遗传相似度GI(对角线之上)和遗传距离GD(对角线之下)
Table 7 Nei’s genetic identity (GI, above diagonal) and genetic distance (GD, below diagonal) among populations
群体 Population SDTA SXXY BJMS SDTA 0.935 5 0.905 9 SXXY 0.066 7 0.934 7 BJMS 0.098 8 0.067 5 表 8 3个白皮松群体方差分析结果
Table 8 ANOVA of the three populations in P. bungeana
变异来源
Source of variation自由度
Degree of freedom (df)离差平方和
Sum of squares (SS)方差成分
Variance component变异所占百分比
Percentage of variation/%群体间 Among populations 2 17.617 0.310 11 群体内 Within population 57 148.450 2.604 89 合计 Total 59 166.067 2.915 100 -
[1] 彭重华, 薄楠林. 白皮松研究进展[J]. 中国农学通报, 2007, 23(11):174−178. doi: 10.3969/j.issn.1000-6850.2007.11.039 Peng Z H, Bo N L. Research progress in Pinus bungeana[J]. Chinese Agricultural Science Bulletin, 2007, 23(11): 174−178. doi: 10.3969/j.issn.1000-6850.2007.11.039
[2] 李斌, 顾万春. 白皮松分布特点与研究进展[J]. 林业科学研究, 2003, 16(2):225−232. doi: 10.3321/j.issn:1001-1498.2003.02.017 Li B, Gu W C. Distribution characteristics and research progress in Pinus bungeana[J]. Forest Research, 2003, 16(2): 225−232. doi: 10.3321/j.issn:1001-1498.2003.02.017
[3] Cai Q, Li B, Lin F, et al. De novo sequencing and assembly analysis of transcriptome in Pinus bungeana Zucc. ex Endl[J]. Forests, 2018, 9(3): 156.
[4] 李斌, 顾万春, 卢宝明. 白皮松天然群体种实性状表型多样性研究[J]. 生物多样性, 2002, 10(2):181−188. doi: 10.3321/j.issn:1005-0094.2002.02.008 Li B, Gu W C, Lu B M. A study on phenotypic diversity of seeds and cones characteristics in Pinus bungeana[J]. Biodiversity Science, 2002, 10(2): 181−188. doi: 10.3321/j.issn:1005-0094.2002.02.008
[5] 王小平, 刘晶岚, 王九龄, 等. 白皮松种子及球果形态特征的地理变异[J]. 北京林业大学学报, 1998, 20(3):28−34. Wang X P, Liu J L, Wang J L, et al. Geographical variation of morphologic characteristics of Pinus bungeana seeds and cones[J]. Journal of Beijing Forestry University, 1998, 20(3): 28−34.
[6] 李斌, 顾万春. 白皮松保育遗传学: 天然群体遗传多样性评价与保护策略[J]. 林业科学, 2005, 41(1):57−64. doi: 10.3321/j.issn:1001-7488.2005.01.011 Li B, Gu W C. Conservation genetics of Pinus bungeana: evaluation and conservation of natural populations’ genetic diversity[J]. Scientia Silvae Sinicae, 2005, 41(1): 57−64. doi: 10.3321/j.issn:1001-7488.2005.01.011
[7] 张雪霞. 基于SCoT标记的白皮松景观基因组学研究[D]. 郑州: 河南农业大学, 2019. Zhang X X. The landscape genomics studies of Pinus bungeana (Pinaceae) based on SCoT molecular markers[D]. Zhengzhou: Henan Agricultural University, 2019.
[8] 刘芳. 基于核基因序列的白皮松谱系地理学研究[D]. 西安: 西北大学, 2011. Liu F. Phylogeography of Pinus bungeana based on nuclear genes[D]. Xi’an: Northwest University, 2011.
[9] 赵罕, 郑勇奇, 李斌, 等. 白皮松天然群体遗传结构的地理变异分析[J]. 植物遗传资源学报, 2013, 14(3):395−401. Zhao H, Zheng Y Q, Li B, et al. Genetic structure analysis of natural populations of Pinus bungeana in different geographical regions[J]. Journal of Plant Genetic Resources, 2013, 14(3): 395−401.
[10] 赵罕, 郑勇奇, 李斌, 等. 白皮松天然群体遗传多样性的EST-SSR分析[J]. 林业科学研究, 2014, 27(4):474−480. Zhao H, Zheng Y Q, Li B, et al. Genetic diversity analysis of Pinus bungeana natural populations with EST-SSR markers[J]. Forest Research, 2014, 27(4): 474−480.
[11] 李斌, 孟庆阳, 李言达, 等. 白皮松种质资源鉴定与评价[J]. 湖南林业科技, 2016, 43(2):1−7. doi: 10.3969/j.issn.1003-5710.2016.02.001 Li B, Meng Q Y, Li Y D, et al. Identification and evaluation of germplasm resources of Pinus bungeana[J]. Hunan Forestry Science Technology, 2016, 43(2): 1−7. doi: 10.3969/j.issn.1003-5710.2016.02.001
[12] 李昕蔓, 金卓颖, 苏安然, 等. 白皮松EST-SSR序列分布特征及引物开发[J]. 林业与生态科学, 2019, 34(3):266−272. Li X M, Jin Z Y, Su A R, et al. EST-SSR sequnence distribution and primer development of Pinus bungeana[J]. Forestry and Ecological Sciences, 2019, 34(3): 266−272.
[13] Kalia R K, Rai M K, Kalia S, et al. Microsatellite markers: an overview of the recent progress in plants[J]. Euphytica, 2011, 177(3): 309−334. doi: 10.1007/s10681-010-0286-9
[14] Ellis J R, Burke J M. EST-SSRs as a resource for population genetic analyses[J]. Heredity, 2007, 99(2): 125−132. doi: 10.1038/sj.hdy.6801001
[15] 张利达, 唐克轩. 植物EST-SSR标记开发及其应用[J]. 基因组学与应用生物学, 2010, 29(3):534−541. Zhang L D, Tang K X. Development of plant EST-SSR markers and its application[J]. Genomics and Applied Biology, 2010, 29(3): 534−541.
[16] 李永强, 李宏伟, 高丽锋, 等. 基于表达序列标签的微卫星标记(EST-SSRs)研究进展[J]. 植物遗传资源学报, 2004, 5(1):91−95. doi: 10.3969/j.issn.1672-1810.2004.01.020 Li Y Q, Li H W, Gao L F, et al. Prograss of simple sequence repeats derived from expressed sequence tags[J]. Journal of Plant Genetic Resources, 2004, 5(1): 91−95. doi: 10.3969/j.issn.1672-1810.2004.01.020
[17] Jamali S H, Cockram J, Hickey L T. Insights into deployment of DNA markers in plant variety protection and registration[J]. Theoretical and Applied Genetics, 2019, 132(7): 1911−1929. doi: 10.1007/s00122-019-03348-7
[18] Rungis D, Bérubé Y, Zhang J, et al. Robust simple sequence repeat markers for spruce (Picea spp.) from expressed sequence tags[J]. Theoretical and Applied Genetics, 2004, 109(6): 1283−1294. doi: 10.1007/s00122-004-1742-5
[19] Bai T D, Xu L A, Xu M, et al. Characterization of masson pine (Pinus massoniana Lamb.) microsatellite DNA by 454 genome shotgun sequencing[J]. Tree Genetics & Genomes, 2014, 10(2): 429−437.
[20] Xiang X Y, Zhang Z X, Wang Z G, et al. Transcriptome sequencing and development of EST-SSR markers in Pinus dabeshanensis, an endangered conifer endemic to China[J]. Molecular Breeding, 2015, 35(8): 1−10.
[21] Echt C S, Saha S, Deemer D L, et al. Microsatellite DNA in genomic survey sequences and unigenes of loblolly pine[J]. Tree Genet Genomes, 2011, 7(4): 773−780.
[22] Liu L, Zhang S J, Lian C L. De novo transcriptome sequencing analysis of cDNA library and large-scale unigene assembly in Japanese red pine (Pinus densiflora)[J]. International Journal of Molecular Sciences, 2015, 16(12): 29047−29059. doi: 10.3390/ijms161226139
[23] Hulce D, Li X, Snyder-Leiby T, et al. Genemarker genotyping software: tools to increase the statistical power of DNA fragment analysis[J]. Journal of Biomolecular Techniques Jbt, 2011, 22(Suppl.): S35.
[24] Kalinowski S T. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment[J]. Molecular Ecology, 2010, 16(5): 1099−1106.
[25] Peakall R, Smouse P E. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research: an update[J]. Bioinformatics, 2012, 28(19): 2537−2539. doi: 10.1093/bioinformatics/bts460
[26] Duan D, Jia Y, Yang J, et al. Comparative transcriptome analysis of male and female conelets and development of microsatellite markers in Pinus bungeana, an endemic conifer in China[J]. Genes, 2017, 8(12): 363.
[27] 周惠娟. 濒危植物白皮松遗传多样性及遗传结构研究[D]. 西安: 西北大学, 2013. Zhou H J. Genetic diversity and population structure of natural endangered forest tree Pinus bungeana in China[D]. Xi’an: Northwest University, 2013.
[28] 杨一欣. 白皮松组植物的群体遗传学和物种形成研究[D]. 西安: 西北大学, 2016. Yang Y X. Population genetics and speciation of Sect. Parrya Mayr[D]. Xi’an: Northwest University, 2016.
[29] 李为民, 李思锋, 黎斌. 利用SSR分子标记分析秦岭冷杉自然居群的遗传多样性[J]. 植物学报, 2012, 47(4):413−421. Li W M, Li S F, Li B. Genetic diversity in natural populations of Abies chensiensis based on nuclear simple sequence repeat markers[J]. Chinese Bulletin of Botany, 2012, 47(4): 413−421.
[30] Tong Y W, Lewis B J, Zhou W M, et al. Genetic diversity and population structure of natural Pinus koraiensis populations[J]. Forests, 2019, 11(1): 39.
[31] Xiang X Y, Zhang Z X, Duan R Y, et al. Genetic diversity and structure of Pinus dabeshanensis revealed by expressed sequence tag-simple sequence repeat (EST-SSR) markers[J]. Biochemical Systematics and Ecology, 2015, 61: 70−77. doi: 10.1016/j.bse.2015.06.001
-
期刊类型引用(3)
1. 吴昊,姜俊慧,罗莉,崔益萌,张婉莹. UV-B辐射增强和氮添加及其互作对空心莲子草-马唐竞争的影响. 西南农业学报. 2024(02): 344-352 .  百度学术
百度学术
2. 吴昊,谢闪闪,苏仪函. 氮添加对茶树与入侵杂草空心莲子草种间竞争的影响. 南方农业学报. 2024(05): 1335-1346 .  百度学术
百度学术
3. 吴昊,刘雨欣,李利军,余梅鹏,杨依琳,庄启国,杜奎. 人为干扰对猕猴桃园杂草多样性与土壤因子关系的影响. 西南农业学报. 2024(06): 1313-1321 .  百度学术
百度学术
其他类型引用(0)



 下载:
下载: