Identification and expression analysis of FmPLT gene family of Fraxinus mandschurica
-
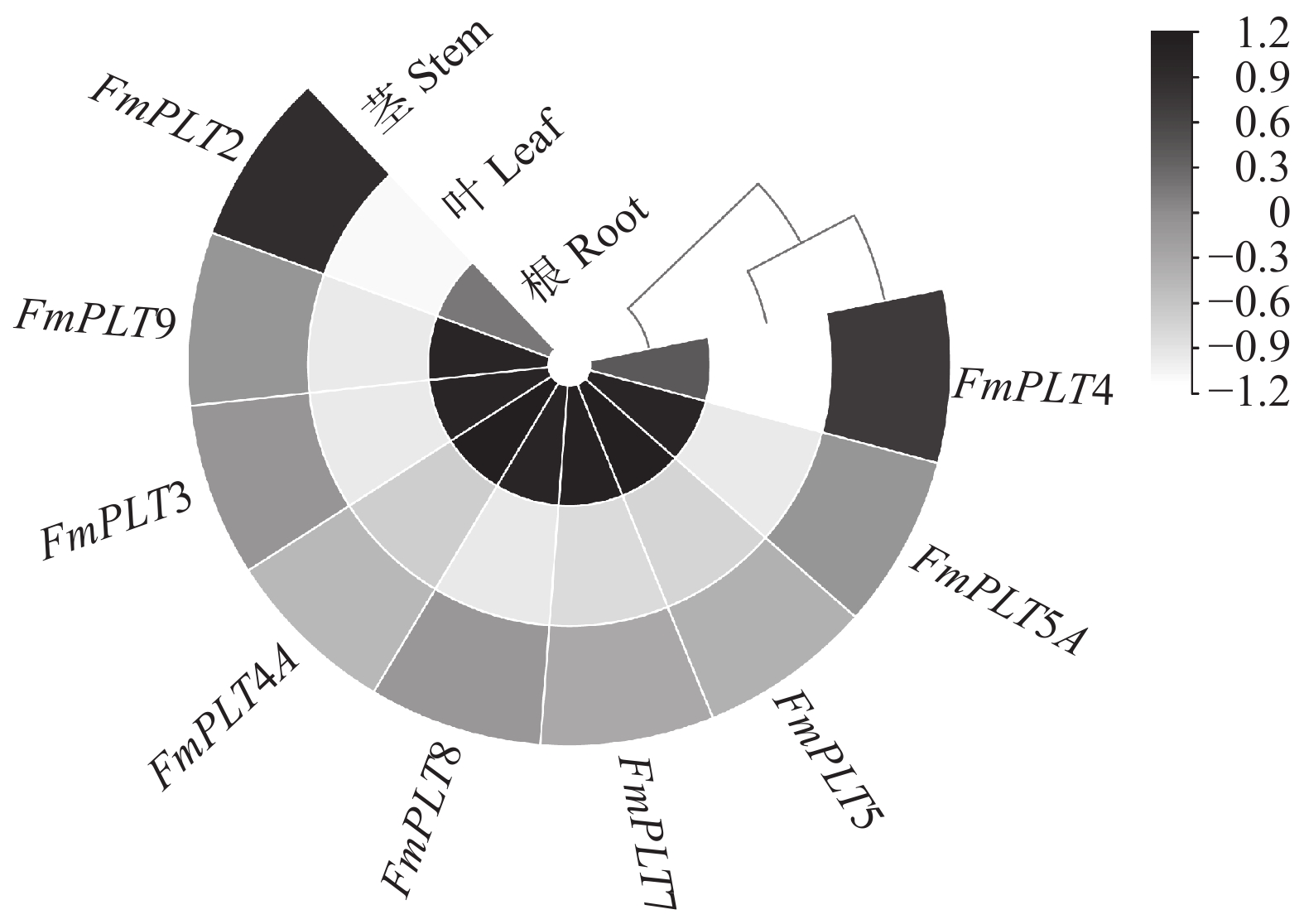
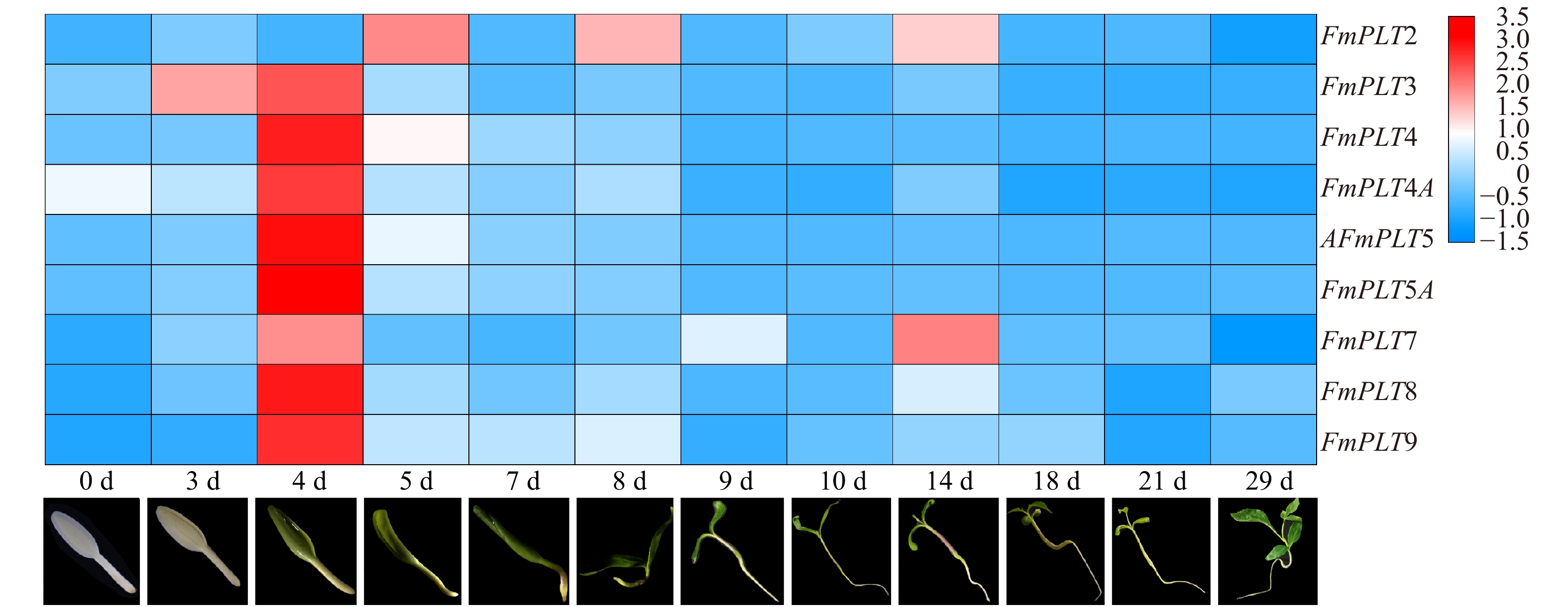
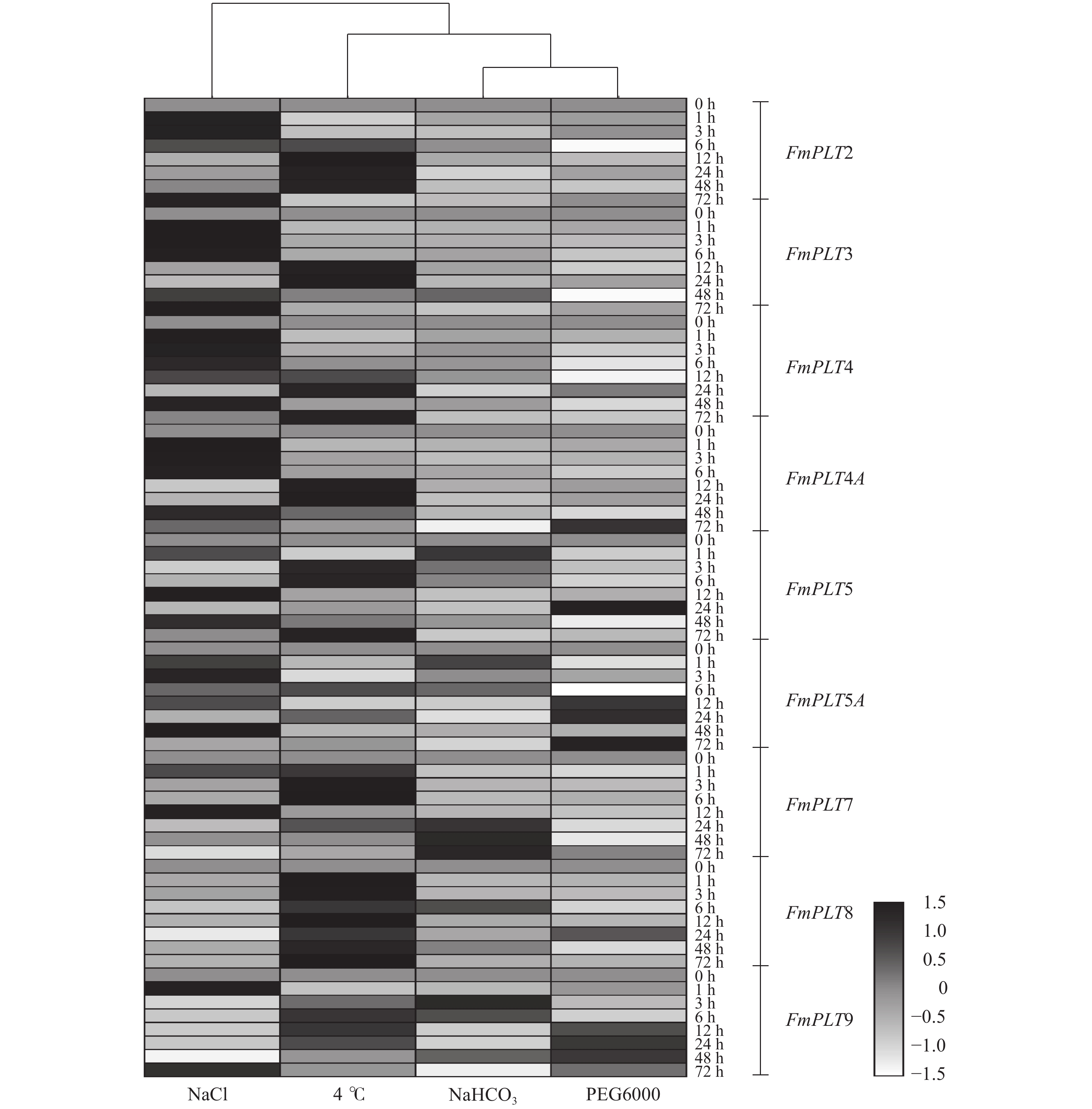
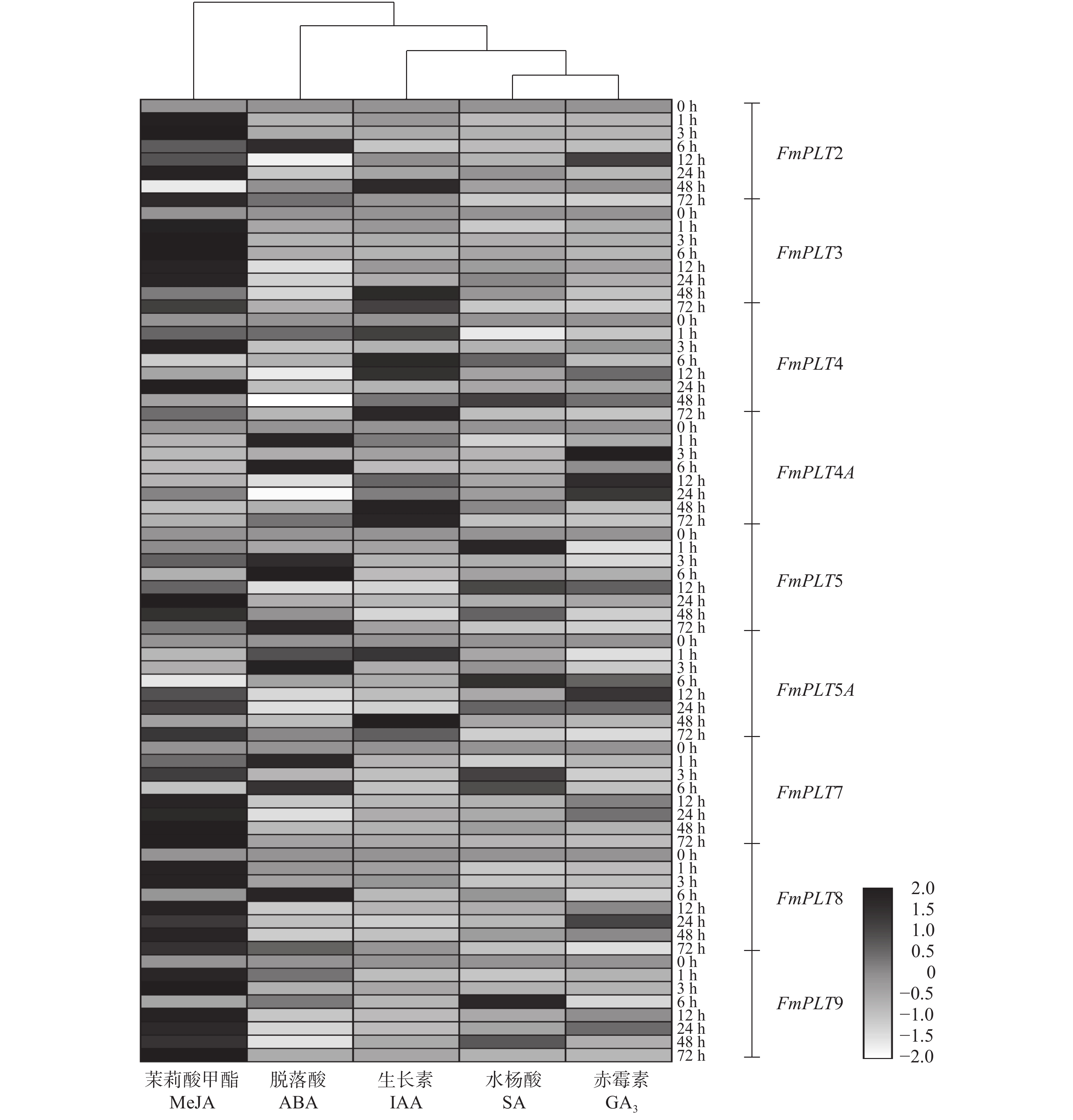
摘要:目的 PLT转录因子家族能够参与植物的再生过程,并且在植物的生长发育及器官建成中发挥着重要作用,本研究旨在探究PLT转录因子在水曲柳生长发育及非生物胁迫中发挥的作用,以期为林木优良基因选育工作提供更多思路。方法 通过同源序列比对和基因克隆等方法对FmPLTs家族的成员进行鉴定及分析,之后对FmPLTs成员进行了详细的生物信息学分析,包括理化性质、系统进化关系和基因结构等;并进一步分析了FmPLTs成员在不同组织及种子萌发过程中的基因表达特征,以及其在非生物胁迫(氯化钠、聚乙二醇6000、碳酸氢钠和低温4 ℃)和植物激素处理(脱落酸、赤霉素、生长素、水杨酸和茉莉酸甲酯下的诱导表达情况。结果 水曲柳基因组中含有9个PLT基因家族成员,分别命名为FmPLT2、FmPLT3、FmPLT4、FmPLT4A、FmPLT5、FmPLT5A、FmPLT7、FmPLT8和FmPLT9。系统进化关系将其分为5组;其在根中表达量最高,在叶中表达量最低;在种子萌发第4 天子叶变绿,下胚轴伸长,此时FmPLTs成员的表达量出现峰值;而在不同激素及非生物胁迫条件下,FmPLTs对低温、干旱、盐胁迫以及MeJA的处理响应明显。结论 FmPLTs参与水曲柳种子萌发及器官发育,并能响应植物激素信号,同时积极参与植物非生物胁迫耐受性的调控,包括低温、高盐和干旱胁迫等;本研究初步揭示了水曲柳FmPLT家族成员响应植物激素和非生物胁迫的表达模式,同时为深入分析FmPLT基因家族基本功能及其调控机制奠定了基础。Abstract:Objective PLT transcription factor family plays an important role in plant growth and organ formation and participates in the regeneration progress. This paper aims to explore the function of PLT transcription factors in the growth and abiotic stress in Fraxinus mandshurica, in order to provide more ideas for forest tree gene selection work.Method By using homologous alignment and cloning techniques, we obtained and identified all members of FmPLT gene family. We conducted detailed bioinformatics analysis of FmPLTs, including physicochemical properties, phylogenetic relationships and gene structure, etc. Besides, we measured the gene expression levels of FmPLTs members in different tissues and seeds during germination, and the expression levels under abiotic stresses (NaCl, PEG6000, NaHCO3, and low temperature 4 ℃) and signal hormone treatmentes (ABA, GA3, IAA, SA and MeJA).Result We revealed that the F. mandshurica genome contained 9 members, which were named as FmPLT2, FmPLT3, FmPLT4, FmPLT4A, FmPLT5, FmPLT5A, FmPLT7, FmPLT8 and FmPLT9, respectively, based on phylogenetic relationship, that could be further divided into 5 groups. The expression of PLT family members was the highest in roots and the lowest in leaves. On the fourth day of seed germination, cotyledons turned green, hypocotyls elongated, most members showed highest expression levels, in response to different hormones and abiotic stresses, FmPLTs showed significant responses to low temperature, drought, salt stress and MeJA treatment.Conclusion FmPLTs are involved in seed germination and organ development of Fraxinus mandshurica, response to plant hormone signaling, and actively participate in the regulation of plant abiotic stress tolerance, including low temperature, high salt and drought stress. This study preliminarily reveals that the expression pattern of FmPLTs in response to both signal hormone treatmentes and abiotic stress, and lays a foundation for in-depth analysis of the basic functions and regulatory mechanisms of FmPLT gene family.
-
Keywords:
- Fraxinus mandschurica /
- PLT transcription factor /
- gene family /
- expression analysis
-
-
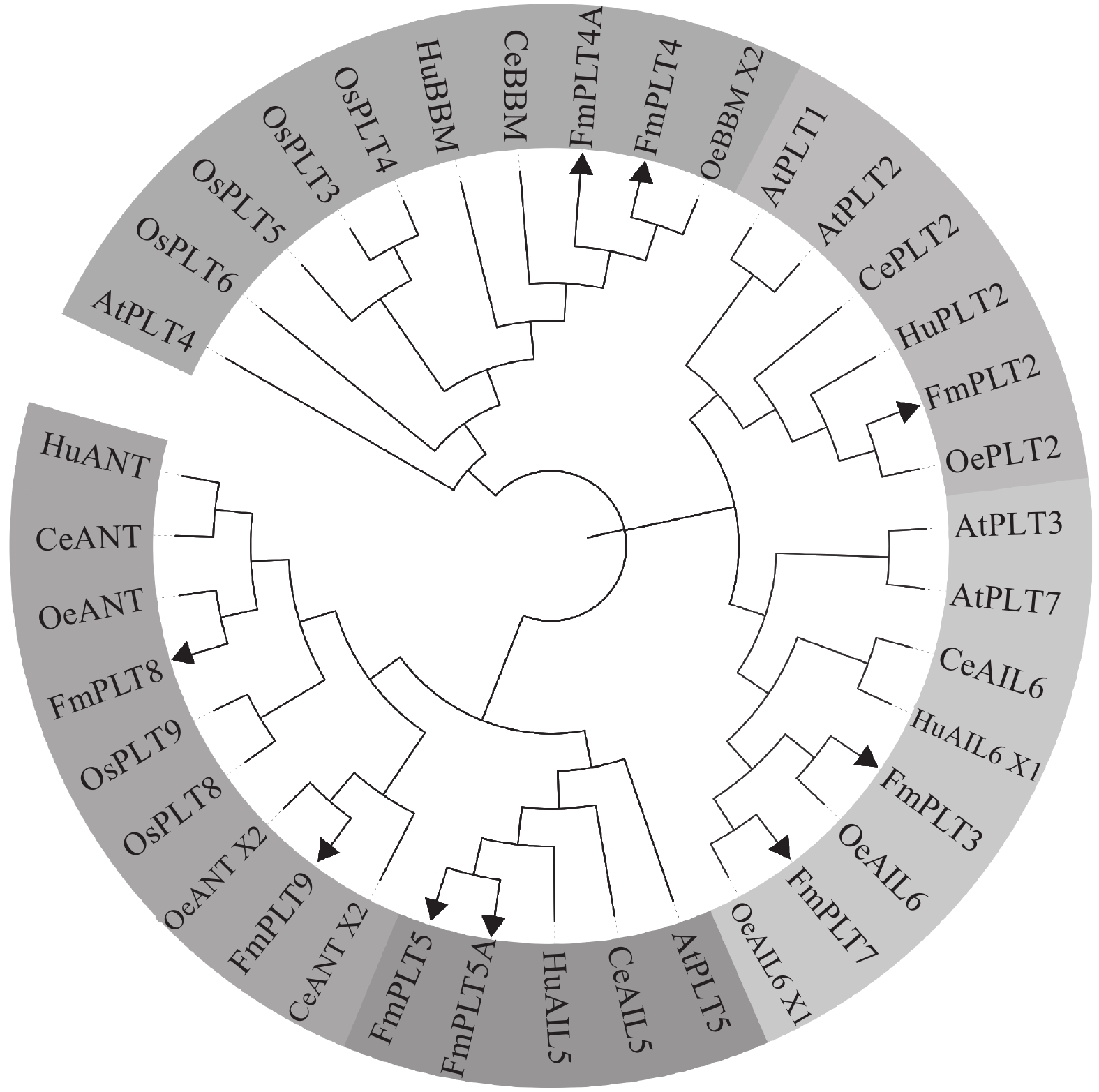
图 1 各物种PLT家族蛋白系统发育进化树
FmPLT2. MW268698;FmPLT3. MW268699;FmPLT4. MW268694;FmPLT4A. MW268695;FmPLT5A. MW268697;FmPLT5. MW268696;FmPLT7. MW268700;FmPLT8. MW268701;FmPLT9. MW268702;AtPLT1. OAP01594.1;AtPLT2. OAP15627.1;AtPLT3. NP_001190279.1;AtPLT4. AAM33803.1;AtPLT5. NP_200549.2;AtPLT7. NP_001318882.1;OsPLT2. AK105909;OsPLT3. AK112070;OsPLT4. AK287726;OsPLT5. AK240892;OsPLT6. AK287621;OsPLT7. AK241712;OsPLT8. AK109848;OsPLT9. AK106306;CePLT2. XP_027164779.1;HuPLT2. XP_021278972.1;OeAIL6. XP_022893842.1;CeAIL6. XP_027179591.1;HuAIL6X1. XP_021275115.1;OeBBMX2. XP_022864559.1;CeBBM. XP_027183775.1;HuBBM. XP_021281898.1;CeAIL5. XP_027175794.1;HuAIL5. XP_021275979.1;OeAIL6X1. XP_022884555.1;OeANT. XP_022870872.1;CeANT. XP_027148114.1;HuANT. XP_021284854.1;OeANTX2. XP_022864341.1;CeANTX2. XP_027178540.1
Figure 1. Phylogenetic tree of PLT family proteins of each species
表 1 FmPLTs基因编码区引物
Table 1 Primers of FmPLTs gene coding region
引物名称 Primer name 引物序列(5′—3′) Primer sequence (5′–3′) PLT2-scence TTGCACATTGAAACTCATCTGAA PLT2-anti-scence GTTCTCCAATCCATTAAGACATCTAA PLT3-scence AGTTGTTTTTTTTTTTCACCGTC PLT3-anti-scence TTAATACGACTGGTGAGGCCAGA PLT4-scence ATGATCAAGAACTGGCTGAGAAATA PLT4-anti-scence GTATACATCCTTAACTTTCCAGTCG PLT4A-scence GCAATAACATCACTGTATCAAGTCAC PLT4A-anti-scence CTACGTATCGTTCCATACCGTAAAA PLT5-scence ATGGATTCTCCTCAGACCTGGC PLT5-anti-scence AAAAGGAAACAAGGCAATAACAAA PLT5A-scence ATGGAAGAAATGAAGAACATGACG PLT5A-anti-scence TCATTCCATTCCGAACATTGG PLT7-scence AAAGAAAAAAAGAAAAAGAAAAATG PLT7-anti-scence CTTTGTGGGTTAGATGAAGGTC PLT8-scence CCCACTTCCCAGCATCTGTT PLT8-anti-scence TTAAGTGTCGTTCCAGGCTGC PLT9-scence TGGAGGGAACCAAACAACACA PLT9-anti-scence GAAAAAAAAACCCTGAAAGTTGAAA 表 2 实时荧光定量PCR引物序列
Table 2 Real-time fluorescence quantitative PCR primer sequences
引物名称 Primer name 引物序列(5′—3′) Primer sequence (5′–3′) qPLT2-scence TGACACTCCGCTTCATAACCA qPLT2-anti-scence AACAGATAATCGGAACCACTCG qPLT3-scence CAGCCGTTCAACCAGTGTCAGTGT qPLT3-anti-scence CCGAGGTTCCTGCATTGCTTGACT qPLT4-scence AATGAAGCACATGACGAGGCAGGAA qPLT4-anti-scence TGCTGGTGGTGTCTTGTGACTCCT qPLT4A-scence GGGAATGCACTTGACAGCCAAACT qPLT4A-anti-scence TCTGCCAGTCCATCTATGCCTTGTT qPLT5-scence CCCATAGGAGGCTTGAACAACA qPLT5-anti-scence ACGGCAGGAGTAGACCACGAAT qPLT5A-scence AGGTGGCAAGCAAGAATCGGAAGG qPLT5A-anti-scence GATTGCCGCAATGTCATAAGCCTCG qPLT7-scence ACAAGAGTCCATTGCCTCGCTAAGA qPLT7-anti-scence CCTTGCTGATGATGCCTCGTAACAC qPLT8-scence ACGGAGTAGGTGAAATGGGTGGTTT qPLT8-anti-scence TGCTGCCTAATGGACTCTTGGTTGT qPLT9-scence AAGAGAACTCCAGGCAGCAGCAA qPLT9-anti-scence TCAGCCATTGTTGGGAGGTGAAGAT TU-scence GCACTGGCCTCCAAGGAT TU-anti-scence TGGGTCGCTCAATGTCAAGG 表 3 氨基酸理化性质分析结果
Table 3 Analysis results of physical and chemical properties of amino acids
基因名
Gene nameGenBank登录号
GenBank accession No.编码区全长
Full length of coding area/bp氨基酸长度
Amino acid length/aa分子量
Molecular mass/Da等电点
Isoelectric point平均亲水性
Average hydrophobicityAP2结构域数量
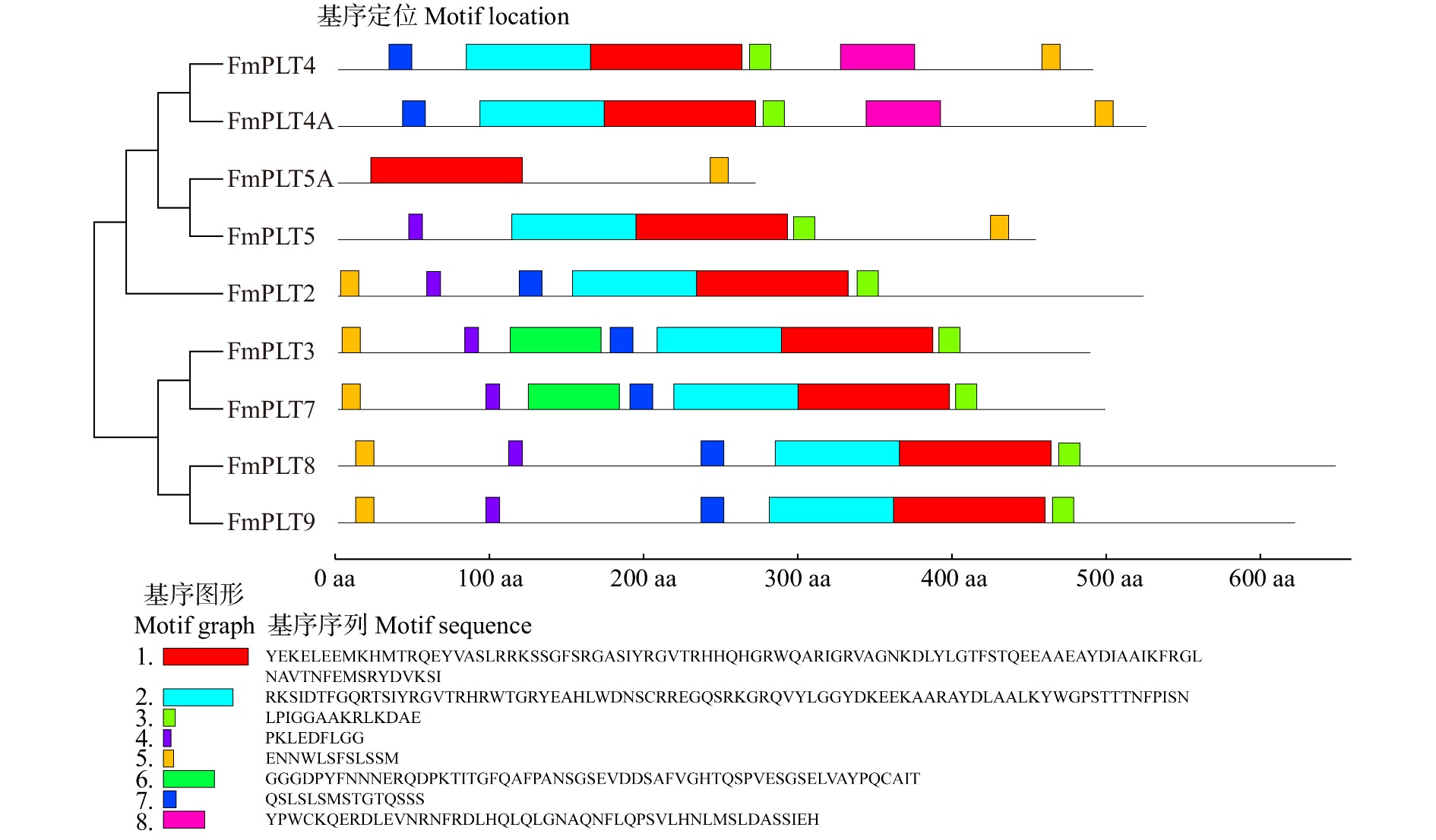
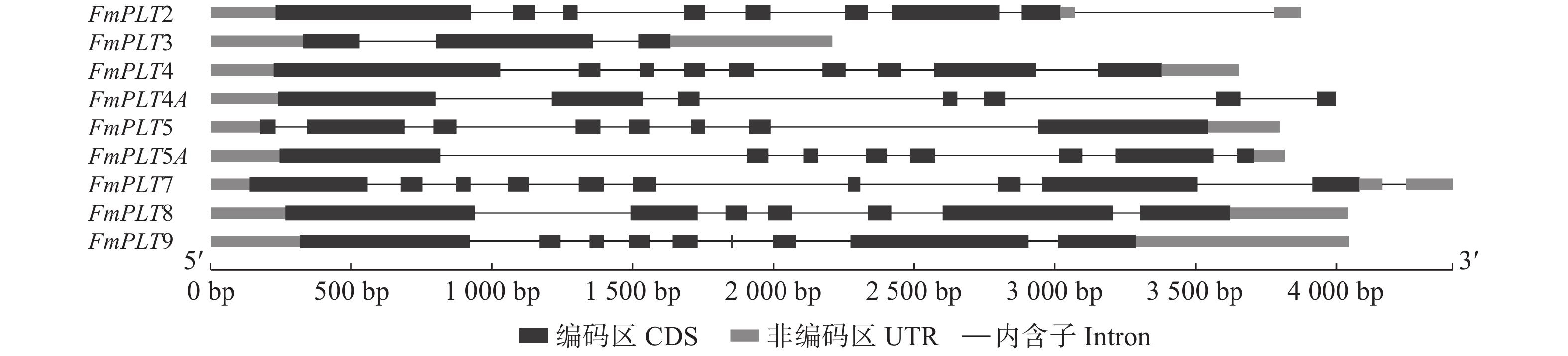
Number of AP2 domainFmPLT2 MW268698 1 599 532 59 045.86 5.89 −0.700 2 FmPLT3 MW268699 1 494 497 55 120.86 5.87 −0.611 2 FmPLT4 MW268694 1 500 499 55 487.44 8.53 −0.705 2 FmPLT4A MW268695 1 605 534 59 122.15 6.68 −0.764 2 FmPLT5 MW268697 1 388 461 50 107.37 7.06 −0.551 2 FmPLT5A MW268696 831 276 30 058.22 7.86 −0.516 1 FmPLT7 MW268700 1 524 507 56 209.03 5.90 −0.612 2 FmPLT8 MW268701 1 980 659 73 471.78 6.62 −0.707 2 FmPLT9 MW268702 1 899 632 70 382.85 6.54 −0.774 2 -
[1] Horstman A, Willemsen V, Boutilier K, et al. AINTEGUMENTA-LIKE proteins: hubs in a plethora of networks[J]. Trends in Plant Science, 2014, 19(3): 146−157. doi: 10.1016/j.tplants.2013.10.010
[2] Aida M, Beis D, Heidstra R, et al. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche[J]. Cell, 2004, 119(1): 109−120. doi: 10.1016/j.cell.2004.09.018
[3] Krizek B A. AINTEGUMENTA utilizes a mode of DNA recognition distinct from that used by proteins containing a single AP2 domain[J]. Nucleic Acids Research, 2003, 31(7): 1859−1868. doi: 10.1093/nar/gkg292
[4] Nole-Wilson S, Krizek B A. DNA binding properties of the Arabidopsis floral development protein AINTEGUMENTA[J]. Nucleic Acids Research, 2000, 21(28): 4076−4082.
[5] Okamuro J K, Caster B, Villarroel R, et al. The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis[J]. Proceedings of the National Academy of Sciences of the United States of America, 1997, 94(13): 7076−7081. doi: 10.1073/pnas.94.13.7076
[6] Berg C V D, Willemsen V, Hendriks G, et al. Short-range control of cell differentiation in the Arabidopsis root meristem[J]. Nature, 1997, 390: 287−289. doi: 10.1038/36856
[7] Akie S, Renze H, Ikram B, et al. Root stem cell niche organizer specification by molecular convergence of PLETHORA and SCARECROW transcription factor modules[J]. Genes, 2018, 32(15−16): 1085−1100. doi: 10.1101/gad.314096.118
[8] Galinha C, Hofhuis H, Luijten M, et al. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development[J]. Nature, 2007, 449: 1053−1057. doi: 10.1038/nature06206
[9] Maehoenen A P, Tusscher K T, Siligato R, et al. PLETHORA gradient formation mechanism separates auxin responses[J]. Nature, 2014, 515: 125−129. doi: 10.1038/nature13663
[10] Blilou I, Xu J, Wildwater M, et al. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots[J]. Nature, 2005, 433: 39−44.
[11] Chen Q, Sun J, Zhai Q, et al. The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis[J]. The Plant Cell, 2011, 23(9): 3335−3352. doi: 10.1105/tpc.111.089870
[12] Mudunkothge J S, Krizek B A. Three Arabidopsis AIL/PLT genes act in combination to regulate shoot apical meristem function[J]. The Plant Journal, 2012, 71(1): 108−121. doi: 10.1111/j.1365-313X.2012.04975.x
[13] Prasad K, Grigg S P, Barkoulas M, et al. Arabidopsis PLETHORA transcription factors control phyllotaxis[J]. Current Biology, 2011, 21(13): 1123−1128. doi: 10.1016/j.cub.2011.05.009
[14] Hofhuis H, Laskowski M, Du Y, et al. Phyllotaxis and rhizotaxis in Arabidopsis are modified by three PLETHORA transcription factors[J]. Current Biology, 2013, 23(11): 956−962. doi: 10.1016/j.cub.2013.04.048
[15] Elliott R C, Betzner A S, Huttner E, et al. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth[J]. The Plant Cell, 1996, 2(8): 155−168.
[16] Bowman J L, Drews G N, Meyerowitz E M. Expression of the Arabidopsis floral homeotic gene Agamous is restricted to specific cell types late in flower development[J]. The Plant Cell, 1991, 3(8): 749−758.
[17] Krizek B A. Ectopic expression of AINTEGUMENTA in Arabidopsis plants results in increased growth of floral organs[J]. Developmental Genetics, 2015, 25(3): 224−236.
[18] Hu L J, Uchiyama K, Shen H L, et al. Nuclear DNA microsatellites reveal genetic variation but a lack of phylogeographical structure in an endangered species, Fraxinus mandshurica, across Northeast China[J]. Annals of Botany, 2008, 102(2): 195−205. doi: 10.1093/aob/mcn074
[19] 林士杰, 张忠辉, 谢朋, 等. 中国水曲柳基因资源的保护与利用[J]. 中国农学通报, 2009, 25(24): 158−162. Lin S J, Zhang Z H, Xie P, et al. Conservation and application of the genetic resource of Fraxinus mandshurica in China[J]. Chinese Agricultural Science Bulletin, 2009, 25(24): 158−162.
[20] Hu L J, Uchiyama K, Shen H L, et al. Multiple-scaled spatial genetic structures of Fraxinus mandshurica over a riparian-mountain landscape in Northeast China[J]. Conservation Genetics, 2010, 11(1): 77−87. doi: 10.1007/s10592-009-0004-0
[21] Zeng F S, Zhou S, Zhan Y G, et al. Drought resistance and DNA methylation of interspecific hybrids between Fraxinus mandshurica and Fraxinus americana[J]. Trees, 2014, 28(6): 1679−1692. doi: 10.1007/s00468-014-1077-z
[22] 卫星. 干旱胁迫对水曲柳苗木细根衰老的影响[D]. 哈尔滨: 东北林业大学, 2008. Wei X. Fine root senescence of manchurian ash seedlings under droughtstress[D]. Harbin: Northeast Forestry University, 2008.
[23] 丁一巍, 詹亚光, 张佳薇, 等. 水曲柳2个PLT转录因子基因的克隆及表达分析[J]. 植物研究, 2019, 39(1): 139−147. doi: 10.7525/j.issn.1673-5102.2019.01.017 Ding Y W, Zhan Y G, Zhang J W, et al. Cloning and expression analysis of two PLT transcription factors genes in Fraxinus mandshurica[J]. Bulletin of Botanical Research, 2019, 39(1): 139−147. doi: 10.7525/j.issn.1673-5102.2019.01.017
[24] Mcphie P. The protein protocols handbook[J]. Analytical Biochemistry, 2003, 315(2): 289.
[25] Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods[J]. Molecular Biology Evolution, 2011, 28(10): 2731–2739. doi: 10.1093/molbev/msr121
[26] Bailey T L, Johnson J, Grant C E, et al. The MEME suite[J]. Nucleic Acids Research, 2015, 43(W1): W39−W49. doi: 10.1093/nar/gkv416
[27] Caffrey D R, Dana P H, Mathur V, et al. PFAAT version 2.0: a tool for editing, annotating, and analyzing multiple sequence alignments[J/OL]. BMC Bioinformatics, 2007, 8(1): 381[2021−01−01]. http://doi.org/10.1186/1471-2105-8-381.
[28] 任小龙, 詹亚光, 梁雪, 等. 水曲柳花发育过程中AG, SOC1基因表达的qRT-PCR分析[J]. 植物研究, 2015, 35(4): 612−617. doi: 10.7525/j.issn.1673-5102.2015.04.021 Ren X L, Zhan Y G, Liang X, et al. QRT-PCR analysis of gene expression of AG and SOC1 during flower development of Fraxinus mandshurica Rupr.[J]. Bulletin of Botanical Research, 2015, 35(4): 612−617. doi: 10.7525/j.issn.1673-5102.2015.04.021
[29] Jofuku K D, Boer B G D, Montagu M V, et al. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2[J]. The Plant Cell, 1994, 9(6): 1211−1225.
[30] Weigel D. The APETALA2 domain is related to a novel type of DNA binding domain[J]. The Plant Cell, 1995, 7(4): 388−389.
[31] Ohme-Takagi M, Shinshi H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element[J]. The Plant Cell, 1995, 2(7): 173−182.
[32] Fujimoto S Y, Ohta M, Usui A, et al. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression[J]. The Plant Cell, 2000, 3(12): 393−404.
[33] Stockinger E J, Gilmour S J, Thomashow M F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit[J]. Proceedings of the National Academy of Sciences of the United States of America, 1997, 3(94): 1035−1040.
[34] Liu Q, Kasuga M, Sakuma Y, et al. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought and low-temperature-responsive gene expression, respectively, in Arabidopsis[J]. The Plant Cell, 1998, 8(10): 1391−1406.
[35] Gilmour S J, Zarka D G, Stockinger E J, et al. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression[J]. The Plant Journal, 1998, 4(16): 433−442.
[36] Kagaya Y, Ohmiya K, Hattori T. RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants[J]. Nucleic Acids Research, 1999, 2(27): 470−478.
[37] Mittler R, Blumwald E. The roles of ROS and ABA in systemic acquired acclimation[J]. The Plant Cell, 2015, 27(1): 64−70. doi: 10.1105/tpc.114.133090
[38] Xue H. Structural characterization and expression pattern analysis of the rice PLT gene family[J]. Acta Biochimica et Biophysica Sinica, 2011, 43(9): 688–697. doi: 10.1093/abbs/gmr068
[39] 徐伟, 严善春. 茉莉酸在植物诱导防御中的作用[J]. 生态学报, 2005, 25(8): 2074−2082. doi: 10.3321/j.issn:1000-0933.2005.08.035 Xu W, Yan S C. The function of Jasmonic acid in induced plant defence[J]. Acta Ecologica Sinica, 2005, 25(8): 2074−2082. doi: 10.3321/j.issn:1000-0933.2005.08.035
-
期刊类型引用(6)
1. 刘树超,邵全琴,杨帆,郭兴健,王东亮,黄海波,汪阳春,刘纪远,樊江文,李愈哲. 黄河源区放牧家畜数量及空间分布无人机遥感调查. 地球信息科学学报. 2021(07): 1286-1295 .  百度学术
百度学术
2. 郑硕,薛兴盛,白杨,吴艳兰. 徐淮地区冬季AOD时空特征及气象因子研究. 环境科学与技术. 2020(01): 78-85 .  百度学术
百度学术
3. 柴曼,周滨,徐威杰,陈晨,刘文权. 基于多尺度分割GF-1影像地表覆盖信息提取研究. 内蒙古煤炭经济. 2020(06): 1-4 .  百度学术
百度学术
4. 贾亮亮,汪小钦,苏华,王峰. 台湾岛高分一号卫星WFV数据气溶胶反演与验证. 环境科学学报. 2018(03): 1117-1127 .  百度学术
百度学术
5. 李裕冬,罗艳,赵海涛,王程亮,杜杰. 基于MaxEnt模型的九寨沟国家级自然保护区川金丝猴适宜生境研究. 四川动物. 2018(05): 585-591 .  百度学术
百度学术
6. 贾亮亮,汪小钦,王峰. 基于波段运算和纹理特征的高分一号多光谱数据云检测. 遥感信息. 2018(05): 62-68 .  百度学术
百度学术
其他类型引用(6)



 下载:
下载: