Screening of internal reference genes of cut flowers of Paeonia lactiflora and expression analysis of key genes of ethylene biosynthesis
-
摘要:目的
筛选芍药切花开放至衰老过程中和盛开期不同组织中稳定表达的最适内参基因,深入解析ACS和ACO基因在乙烯介导的芍药切花开放至衰老过程中的功能。
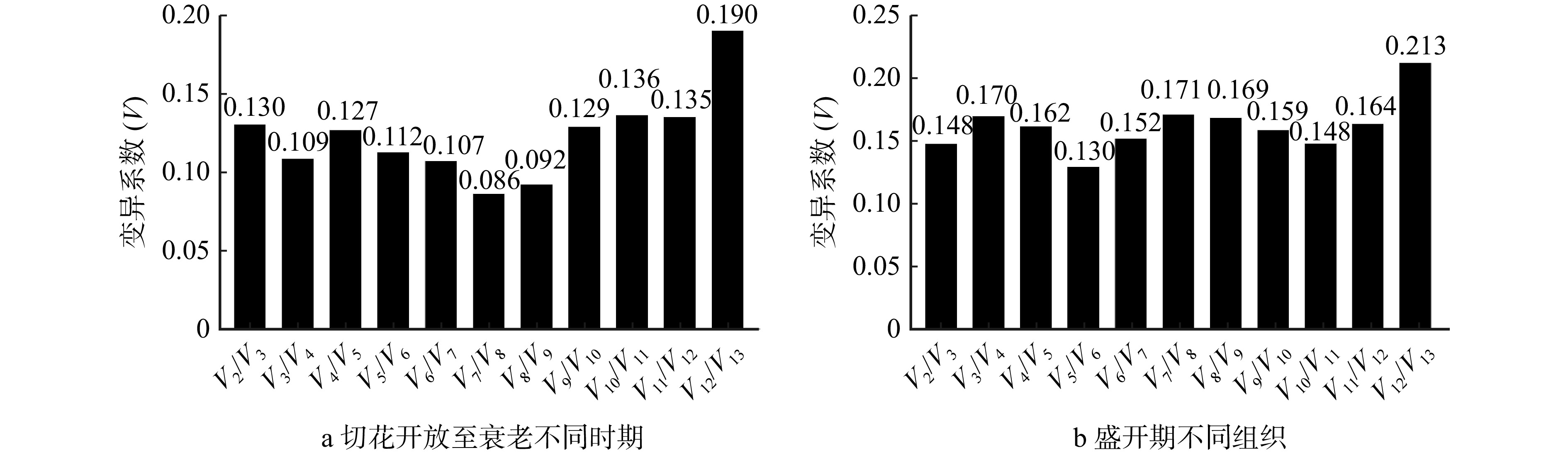
方法(1)以芍药‘桃花飞雪’为材料,基于qRT-PCR技术分析13个候选内参基因在芍药切花开放至衰老不同时期及盛开期不同组织中表达水平的变化,应用geNorm、NormFinder、BestKeeper等软件和ΔCt法对其表达稳定性进行了评价;通过在线RefFinder网站分析获得候选内参基因表达稳定性的综合排名。(2)利用筛选出的双内参基因,对乙烯生物合成关键基因ACS和ACO在芍药切花开放至衰老不同时期及盛开期不同组织中的表达水平进行定量分析。
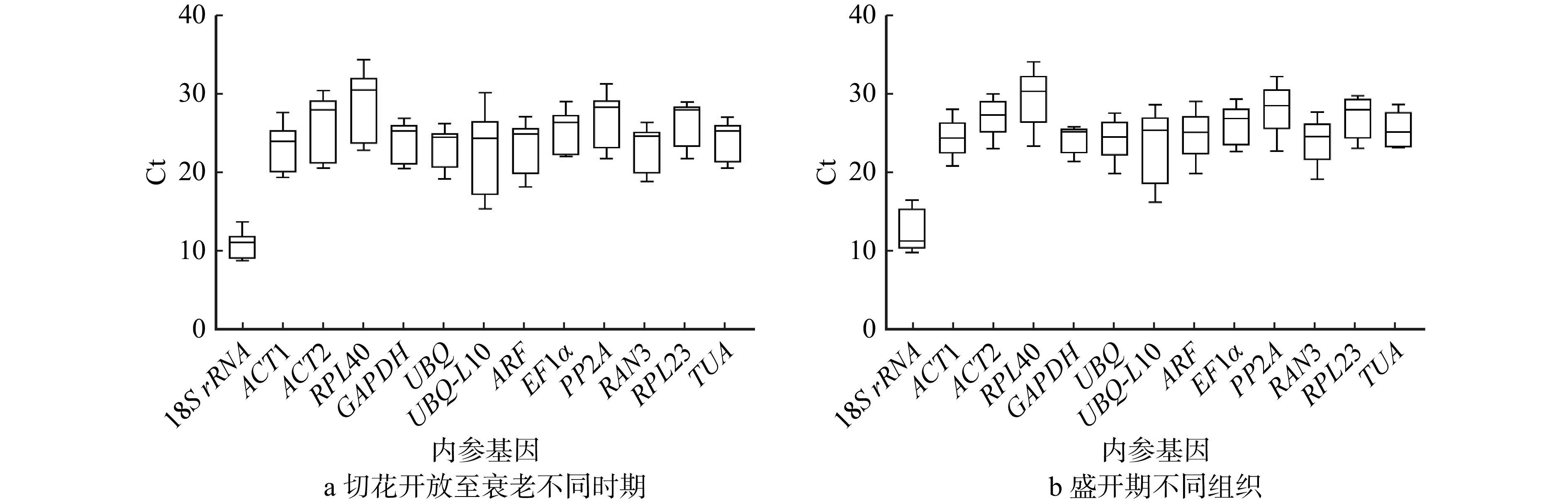
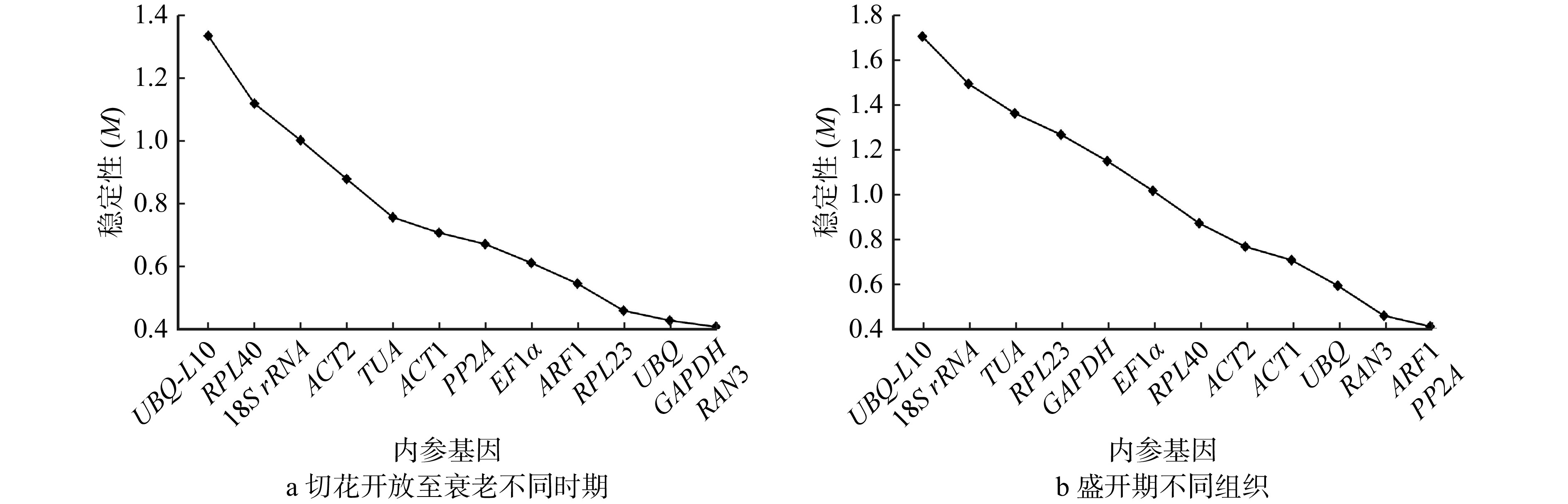
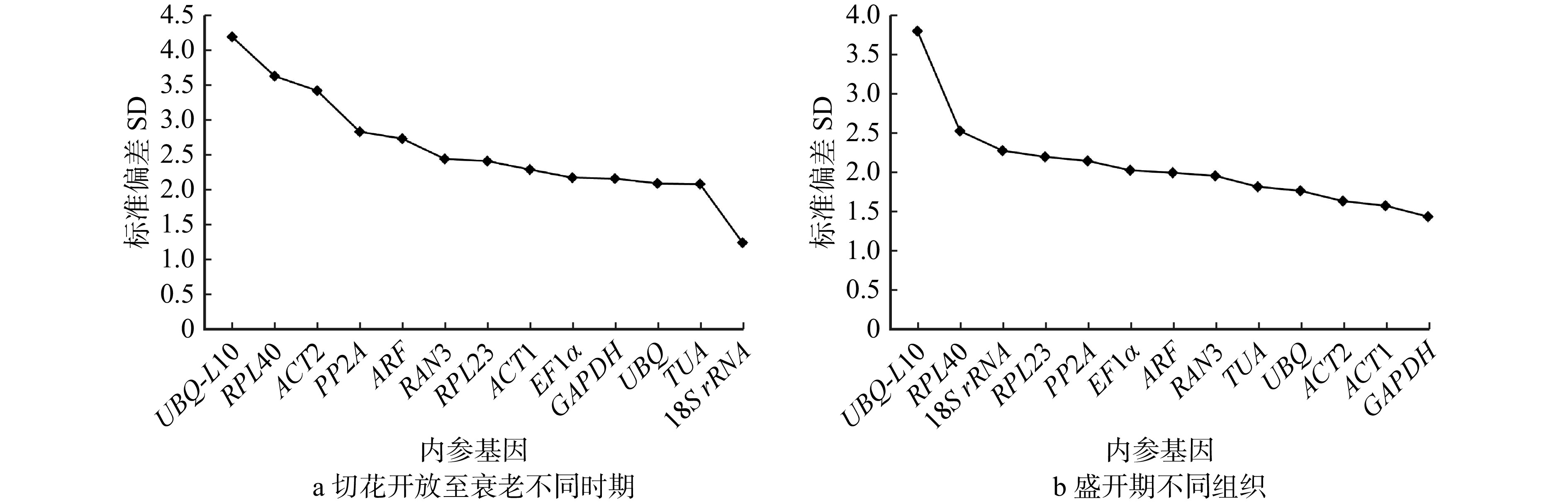
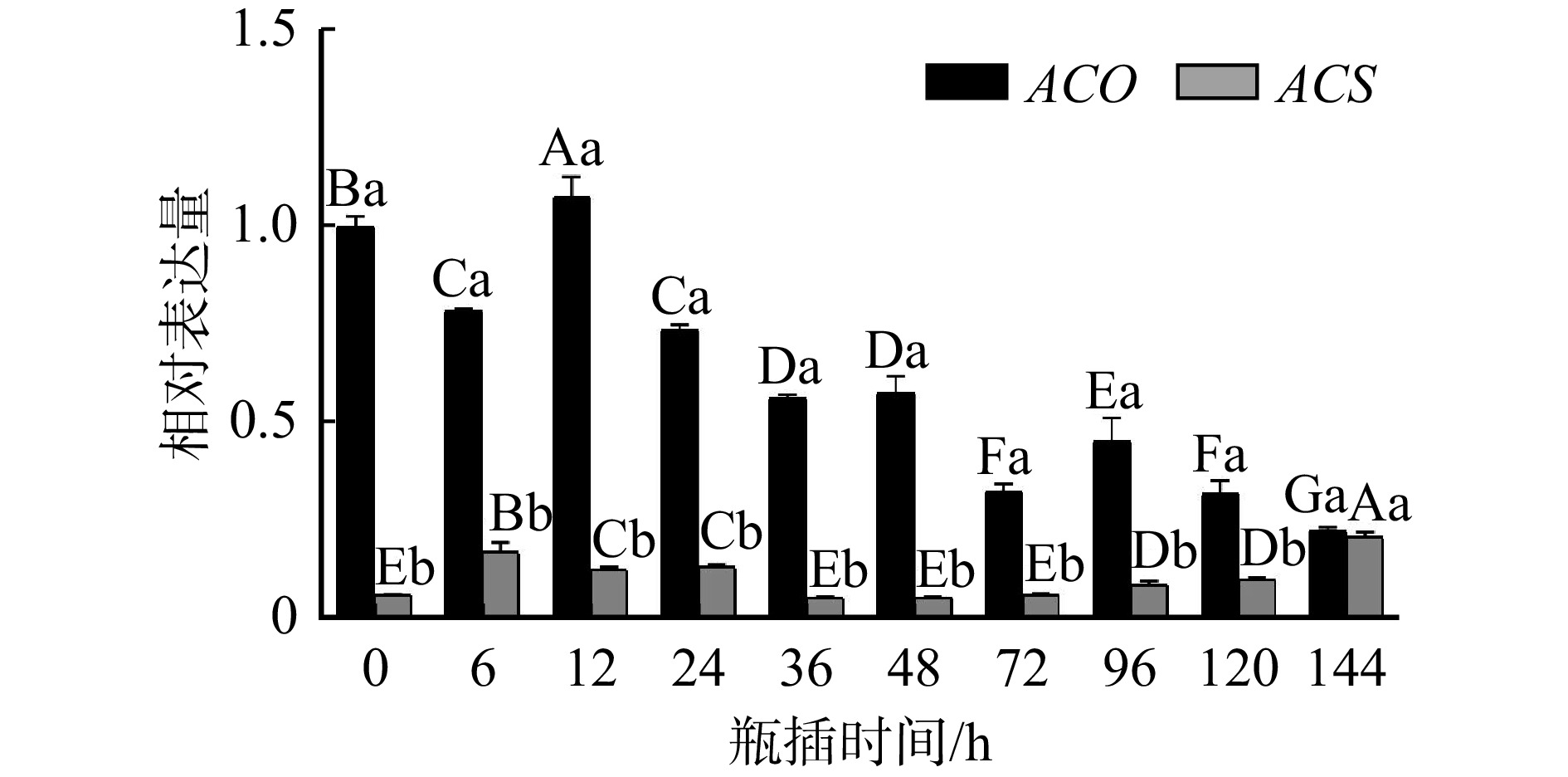
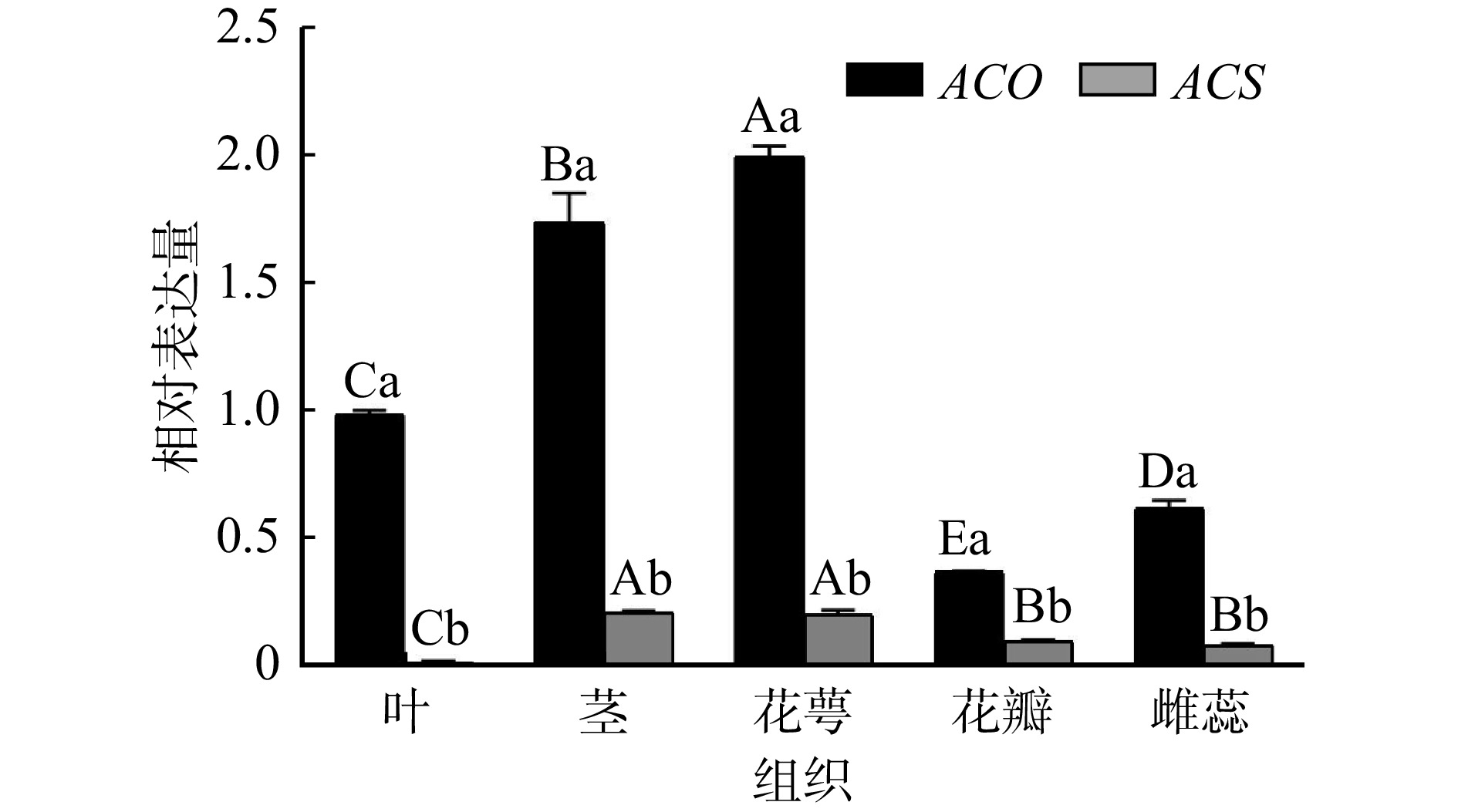
结果(1)在芍药切花开放至衰老的不同时期,geNorm分析表明GAPDH和RAN3表达稳定性最好;NormFinder分析显示RAN3表达最稳定;BestKeeper分析表明18S rRNA表达稳定性最好;RefFinder综合分析表明RAN3和GAPDH为最适内参基因。(2)在盛开期芍药切花花器官不同组织中,geNorm分析表明ARF和PP2A表达稳定性最好;NormFinder分析显示UBQ表达最稳定;BestKeeper分析表明GAPDH表达稳定性最好;RefFinder综合分析表明UBQ和ARF为最适内参基因。(3)在芍药切花开放至衰老过程中,ACS基因表达量显著低于ACO基因表达量,是调控乙烯合成的开关;ACO主要在切花开放前期(瓶插0 ~ 12 h)发挥作用;ACS表达量在绽口期和衰败期(瓶插6 h和144 h)均发挥重要作用,并在盛开期对乙烯合成起到限制作用;(4)在芍药切花盛开期不同组织中,ACS基因表达量显著低于ACO,ACS和ACO基因的表达存在组织差异性,且均在茎和花萼中表达较高。
结论本研究筛选出芍药切花开放至衰老不同时期(RAN3和GAPDH)以及盛开期不同组织中(UBQ和ARF)qPCR定量表达分析的双内参基因组合;乙烯生物合成关键基因ACO主要在芍药切花开放过程发挥作用,ACS基因在芍药切花开放和衰老过程均发挥重要的作用。研究结果将为切花寿命的改良提供参考。
Abstract:ObjectiveThis study aims to select the most suitable reference genes, which are stably expressed during the process from the opening to senescence and different tissues in blooming period of peony cut flowers, and to deeply analyze the function of ACS and ACO genes in ethylene-mediated process of peony cut flower opening to senescence.
Method(1) With cultivar ‘Taohuafeixue’ as materials, the variations of expression levels of 13 candidate internal reference genes at different stages from blooming to senescence and in different tissues of the full bloom stage in herbaceous peony cut flowers were analyzed by qRT-PCR. The expression stability of 13 candidate internal reference genes was assessed by ΔCt and 3 softwares including geNorm, NormFinder, and BestKeeper. The comprehensive rankings of expression stability of candidate internal reference genes were obtained by online RefFinder analysis. (2) Based on the selected double internal reference genes, the expression profile of ACS and ACO, key genes of ethylene biosynthesis were quantitatively analyzed at different stages from blooming to senescence and in different tissues of full bloom stage in herbaceous peony cut flowers.
Result(1) geNorm analysis showed that the expression stability of GAPDH and RAN3 at different stages was best. NormFinder analysis indicated that the expression stability of RAN3 was optimal. BestKeeper analysis illustrated that 18S rRNA expression was the most stable. RefFinder analysis indicated that RAN3 and GAPDH were the best suitable internal reference genes at different stages. (2) geNorm analysis indicated that the expression of ARF and PP2A in different tissues had the best expression stability. NormFinder analysis showed that the expression of UBQ was the most stable. BestKeeper analysis illustrated that GAPDH was best. RefFinder analysis indicated that UBQ and ARF were the best internal reference genes in different tissues in herbaceous peony cut flowers. (3) From opening to senescence process of peony cut flowers, the expression of ACS gene was significantly lower than that of ACO gene, and ACS was the ‘switch’ for the regulation of ethylene synthesis. ACO mainly played a role in pre-opening stage of cut flowers (0−12 h). ACS expression played an important role both in the bloom stage and in the senescence stage (6 h and 144 h), and played a limiting role in ethylene synthesis during the bloom stage. (4) ACS expression was significantly lower than ACO in different tissues of peony cut flowers during blooming period, there was tissue variability in the expression of ACS and ACO, and both of them were higher in stems and calyxes.
ConclusionThis study screens double internal reference gene combinations for qPCR quantitative expression analysis in different tissues of herbaceous peony cut flowers at varied periods from opening to senescence (RAN3, GAPDH) and at full bloom (UBQ, ARF). ACO gene, a key gene of ethylene biosynthesis mainly plays a role in the blooming process, and ACS plays an important role in both blooming and senescence process in herbaceous peony cut flowers. The finding will provide a reference for the improvement of cut flower longevity.
-
近年来,木材被广泛应用于室外领域,如木结构建筑、木栈道、木围栏等。然而在室外应用时,木材会不可避免地受到自然环境因素的影响,产生腐朽等生物劣化现象[1],不仅缩短了其使用寿命,还会造成安全隐患。目前,户外木材主要采用南方松(Pinus spp.)和欧洲赤松(Pinus sylvestris)等松木为原料。并且有研究表明:相比于采绒革盖菌(Coriolus versicolor)等白腐菌,松木等针叶材更易受密黏褶菌(Gloeophyllum trabeum)和绵腐卧孔菌(Poria vaporaria)等褐腐菌侵害,且褐腐菌能够在较短时间内快速降解木材,在质量损失较低的情况下导致木材力学强度急剧下降[2],严重影响其使用价值。因此,阐明木材在褐腐初期的微观结构和化学成分变化对于木材防腐保护具有重要意义。
在褐腐过程中,轴向排列的细胞有利于真菌沿木材的顺纹方向蔓延生长,但实际应用中,木材通常要经过封端处理以防止端裂、腐朽等劣化发生。而对于花纹美观且直接暴露的弦切面与径切面,菌丝进入木材内部的通道主要为射线薄壁组织、细胞壁纹孔等[3-5]。随着褐腐的进行,木材中的纤维素和半纤维素被陆续降解,而木质素基本不被破坏[6],因此残留的木质素使得木材在宏观上通常呈现出红褐色[7]。研究表明:在褐腐过程中半纤维素首先发生降解,其降解速度比纤维素更快[8-9]。此外,腐朽材中纤维素的结晶度也明显降低,有研究显示:褐腐15周后的马尾松(Pinus massoniana)相对结晶度下降了60.05%[10],这表明结晶纤维素在褐腐过程中遭到破坏,原本排列有序的分子链被打乱,分子间作用力减小,进而导致分子间间隙增加。褐腐初期对于木材性能的影响非常显著。Witomski等[11]利用粉孢革菌(Coniophora puteana)对欧洲赤松进行腐朽试验,发现褐腐初期纤维素的聚合度由6 000降至1 800,而此时的质量损失仅为7%。尽管褐腐初期木材的质量损失较低(通常不超过10%[12]),但会使木材力学强度急剧下降[13]。
综上所示,以往研究的褐腐周期一般较长(12周),且大多关注腐朽带来的最终结果。对腐朽各阶段,尤其是褐腐初期,木材组分及宏、微观变化的研究并不深入。因此,本研究对户外常用的南方松边材进行不同时长的褐腐处理,重点关注腐朽初期木材的各项变化,揭示褐腐菌进入木材内部的通道,并阐明其对木材微观结构和化学成分变化的影响,为深入探究木材褐腐机理奠定理论基础。
1. 材料与方法
1.1 材 料
南方松边材,试件尺寸为10 mm (轴向) × 20 mm (弦向) × 20 mm (径向);饲木选用南方松边材,尺寸为3 mm(轴向) × 20 mm (弦向) × 20 mm(径向)。褐腐菌采用密黏褶菌,购自中国普通微生物菌种保藏管理中心。
1.2 褐腐试验
参照GB/T 13942.1—2009 《木材耐久性能第一部分:天然耐腐性实验室试验方法》[14]进行土壤木块法测试,腐朽时长分别为0、10、20、40 d。试件在腐朽过程中的质量损失率(L)按公式(1)计算:
L=m0−m1m0×100% (1) 式中:L为试件质量损失率,%;m0为试件腐朽前的绝干质量,g;m1为试件腐朽后的绝干质量,g。
1.3 颜色测定
利用色差仪(三恩施NH310,中国)对木材腐朽前后弦切面的颜色进行表征,测得CIE色度系统中的参数L*、a*和b*。L*为明度值(白色为100,黑色为0),a*为红绿色品指数(a*值越大,颜色越偏红,反之偏绿),b*为黄蓝色品指数(b*值越大,颜色越偏黄,反之偏蓝)。每块试件选取5个点位进行测试,并计算平均值。腐朽过程中的总色差(ΔE)按式(2)计算:
ΔE=√ΔL∗2+Δa∗2+Δb∗2 (2) 式中:ΔE为腐朽前后木材的总色差;ΔL*、Δa*、Δb*分别为不同腐朽时间后腐朽材与健康材的L*、a*、b*差值。
1.4 化学成分测定
试件的苯−乙醇抽提物、酸不溶木质素、综纤维素、纤维素含量,分别根据GB/T 2677.6—94《造纸原料有机溶剂抽出物含量的测定》[15]、ASTM D 1106—96 “Standard Test Method for Acid-Insoluble Lignin in Wood”[16]、Browning(1967)的综纤维素改进测定法[17]、硝酸−乙醇纤维素测定法[18]进行测试。半纤维素含量由综纤维素与纤维素含量之差得到。
1.5 微观形貌表征
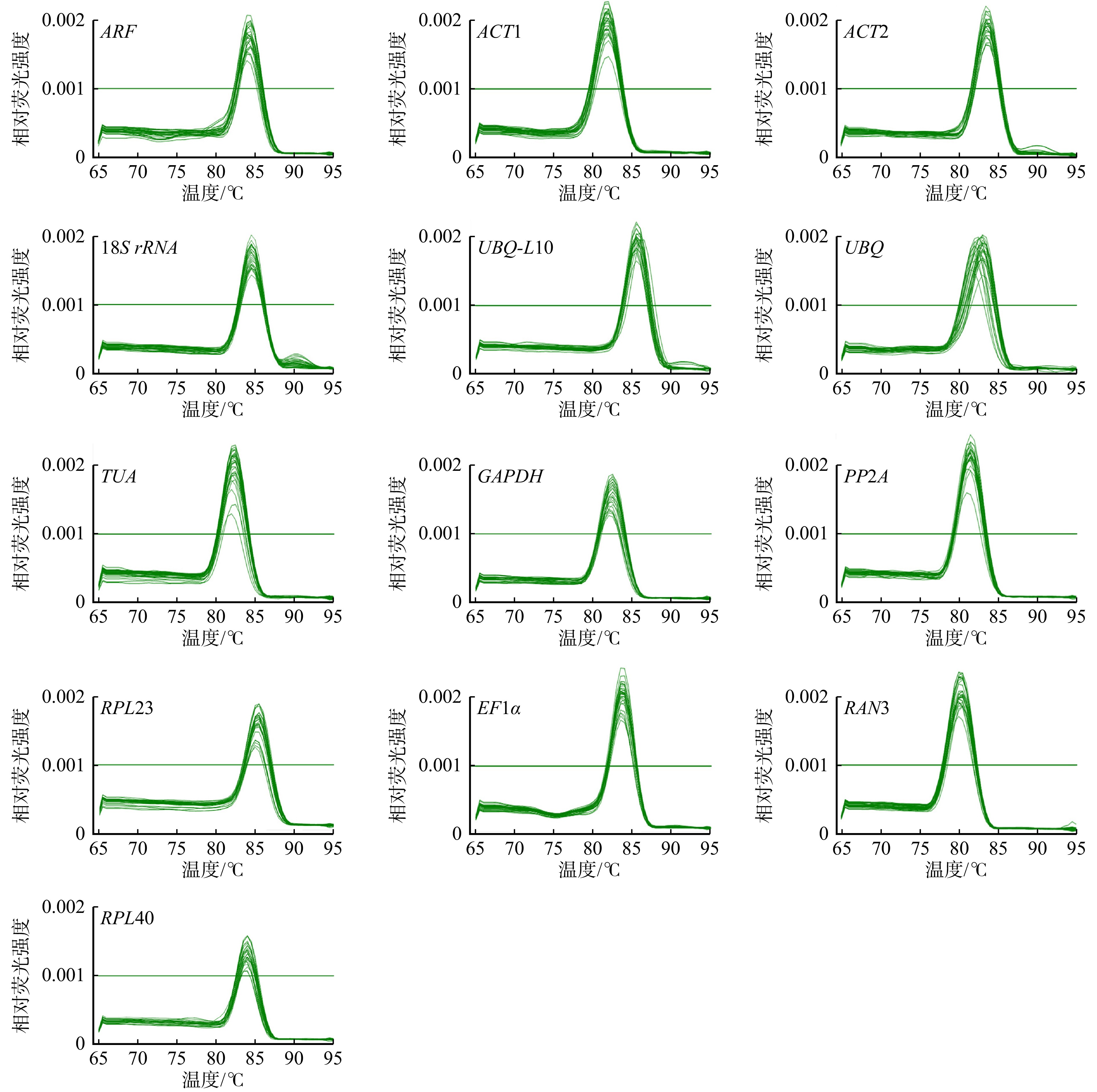
收集不同腐朽时长的试件,并在其弦切面与横切面上分别制取5 mm × 5 mm薄片,利用场发射扫描电子显微镜(FE-SEM,日立SU8010,日本)进行观察。同时,使用ImageJ软件测量木材在腐朽过程中细胞壁厚度的变化。
1.6 红外光谱表征
利用傅里叶红外光谱仪(FTIR,Nicolet IS 10,美国),通过KBr法测定试件的红外光谱,扫描范围为400 ~ 4 000 cm−1,扫描次数为64次,分辨率为4 cm−1。
1.7 相对结晶度测定
利用X射线衍射仪(XRD,Bruker D8 ADVANCE,德国)、Jade 6.0软件对试件进行测试与分析。扫描角度范围为5° ~ 40°(2θ),扫描速率为2.0°/min,步长0.02°。
根据Scherrer公式计算微晶尺寸[19]:
Cs=Kλβcosθ (3) 式中:Cs为微晶尺寸,Å;K为校正系数,取0.90;λ为X射线衍射波长,取1.54 Å;β为衍射峰的半高宽,°;θ为布拉格角,°。
根据Segal公式计算相对结晶度[20]:
Cr=I200−IamI200×100% (4) 式中:Cr为相对结晶度,%;I200为(200)晶格衍射角的总强度,2θ = 22.4°,即结晶区的衍射强度;Iam为(110)与(200)晶格之间最小强度,即非结晶区衍射的散射强度,2θ = 18.4°。
2. 结果与讨论
2.1 宏观颜色变化分析
由图1可知:经褐腐菌侵染后,木材的表面(弦切面)颜色发生明显变化,从原来的偏黄色变为灰褐色。随着腐朽的进行,木材表面的ΔL*值持续降低,表明木材颜色变暗(图1b)。同时,Δa*与Δb*值总体呈增加趋势,表明腐朽后木材表面更偏向红褐色。随着腐朽程度的深入,木材中的综纤维素被大量脱除,残留的木质素使木材呈现为红褐色,色差值进一步增大。
2.2 微观形貌变化分析
图2和图3分别为南方松边材在腐朽不同时长后的弦切面和横切面电镜照片。在此过程中,木材的质量变化和细胞壁厚度变化情况如图4所示。由图2可知:未经腐朽的试件显示出较为光滑平整的弦切面(图2a),然而其横切面表面(图3a)还残留着一些破碎的木材组织,这主要由试件的锯切加工过程导致。腐朽10 d后,这些残留的木材组织被逐步降解,在横切面上裸露出木材的细胞腔与细胞壁(图3b)。同时,对于径向排列的射线薄壁细胞,可以观察到其内部菌丝已经穿透细胞壁(矩形框线内的截取图像),并横穿细胞腔,有延伸到下一个细胞的趋势。此外,在弦切面上(图2b)可以发现,木材表面被菌丝附着,同时细胞壁上部分具缘纹孔的纹孔膜被降解并发生破裂(矩形框内的放大图像),菌丝穿透纹孔进入木材细胞腔。研究表明,纹孔膜的主要成分为半纤维素与少量纤维素[21],为褐腐菌降解木材的主要成分。褐腐10 d后,木材内部残留的菌丝较少,结合图4可知,此时的木材质量损失率较低,仅为2.77%。腐朽20 d后,木材的质量损失率增大为16.60%,表明褐腐菌的生长迅速,对营养物质的代谢更剧烈,加快了对木材的降解进程。此时,在木材的管胞内(图2c、图3c)观察到大量交叉缠绕的菌丝,部分菌丝正从纹孔处进入细胞腔(图2c箭头位置),并在细胞腔内蔓延生长,表明褐腐菌逐步完成初期定植。此外,从横切面上可以观察到,木材的S2层被严重降解,细胞壁厚度损失率高达18.24%(图4b)。随着腐朽天数的延长,菌丝的数量不断增加,木材的质量和细胞壁厚度进一步降低。腐朽40 d后,木材的弦切面基本被菌丝覆盖(图2d),而横切面上的木材细胞壁也不再完整,由于纤维素的降解,细胞壁结构逐渐失去支撑作用,出现溃烂瓦解的现象(图3d)。此时,木材的质量损失率和细胞壁厚度损失率分别为20.35%和20.86%(图4),相比于之前,木材的降解速度有所减缓,据此推测腐朽20 d时菌丝已基本完成初期定植。
2.3 化学成分变化分析
腐朽过程中,木材中各组分的变化如表1所示,其对应的FTIR谱图如图5所示。由图5可知:相比于健康材,腐朽10 d后,木材中各特征峰的强度变化较小,质量损失率较低(仅为2.77%),表明褐腐初期木材的降解速度缓慢。由表1可知:此时的质量损失主要来源于抽提物和半纤维素含量的减少,两者的质量损失率分别为47.55%和49.19%。木材中抽提物的绝对含量很少,且成分复杂,除能够被腐朽菌利用外,部分还具有抑菌作用[22],因此其在褐腐初期的变化还有待进一步探讨。由此推测,在腐朽初期,褐腐菌主要降解木材中的半纤维素。随着腐朽时间的延长,木材中综纤维素相对质量分数不断降低,而木质素的相对质量分数有所增加。褐腐20 d时,腐朽材在1 736 cm−1(半纤维素中的乙酰基和羰基的C=O伸缩振动)、1 372 cm−1(纤维素中的C—H变形振动)、897 cm−1(纤维素中的C—H变形振动)和810 cm−1(半纤维素中的葡甘露聚糖)[23-26]处的峰强开始明显降低,表明木材中的碳水化合物发生了严重的降解。碳源作为营养物质被真菌代谢,以及大分子解聚导致3 342 cm−1(纤维素中的O—H伸缩振动)和2 860 cm−1(对称CH2的伸缩振动)[27]处的峰强增加。此时,半纤维素的质量损失率高达85.88%,而纤维素质量分数仅下降了3.54%。相反,木质素特征峰的强度显著增加,如1 510 cm−1(芳环的C=C骨架振动)、1 225 cm−1(C—O伸缩振动)处[23-26],此时木质素相对质量分数增加了16.07%。
表 1 不同腐朽时间后木材的质量损失及化学成分变化Table 1. Mass loss and chemical composition of wood samples at different decay times腐朽时间
Decay
time/d质量损失率
Mass loss
rate/%抽提物质量分数
Extract mass
fraction/%木质素质量分数
Lignin mass
fraction/%综纤维素质量分数
Holocellulose mass
fraction/%纤维素质量分数
Cellulose mass
fraction/%半纤维素质量分数
Hemicellulose mass
fraction/%0 0 3.26 28.07 68.67 50.05 18.62 10 2.77 1.71 28.11 60.12 50.66 9.46 20 16.60 2.77 31.29 50.91 48.28 2.63 40 20.35 3.04 32.58 48.91 46.68 2.23 综上可知,腐朽10 ~ 20 d内是褐腐菌定植木材的重要阶段,此时木材的质量急剧降低,其中的半纤维素和纤维素被迅速降解,细胞壁和纹孔的结构发生破坏,为褐腐菌深入木材进行后续降解奠定了基础。
2.4 相对结晶度分析
由化学成分变化分析可知,褐腐初期半纤维素的降解优先于纤维素,且降解程度更加剧烈。尽管纤维素在这一过程中的损失较少,但其结构也发生了不同程度的变化。本研究对腐朽不同时长后,木材中纤维素的晶格间距d200、微晶尺寸Cs、相对结晶度Cr变化进行了表征,结果如表2所示。总体而言,各阶段的腐朽材的(200)晶面均位于22.4°附近(介于22.30° ~ 22.45°之间),说明腐朽过程对纤维素结晶区的影响相对较小。相比于健康材,腐朽材的晶格间距减小,这主要是因为纤维素结晶区外部松散的非晶区域或不完全结晶的物质被脱除,导致剩余的结晶区更加有序地排列[28]。褐腐20 d后,由于半纤维素含量急剧降低,结晶区在氢键作用下紧密靠拢,因而此时晶格间距d200最小(3.962 Å),相对结晶度Cr从原来的38.63%增加到47.02%。结晶度的增加及晶格间距的减小将阻碍褐腐菌的代谢产物渗透进入纤维素结晶区,因此20 d后木材的腐朽降解速率变缓。然而,随着半纤维素的大量脱除,木材中的孔隙结构增多,褐腐菌将以酶降解的方式进一步对木材细胞壁进行破坏[29]。因此,腐朽40 d后,褐腐菌对半纤维素的降解速度减缓,逐步开始降解纤维素,因而导致其相对结晶度有所降低(降低为44.21%),晶格间距逐渐变大(3.972 Å)。此外,在腐朽过程中,由于微纤丝的不断聚集,使得其微晶尺寸逐渐增加。
表 2 不同腐朽时间后木材的微晶尺寸和相对结晶度变化Table 2. Changes in crystallite sizes and relative crystallinity of wood samples at different decay times腐朽时间
Decay
time/d2θ/(°) 晶格间距
Lattice distance
(d200)/Å微晶尺寸
Crystallite size
(Cs)/Å相对结晶度
Relative crystallinity
(Cr)/%0 22.31 3.982 75.29 38.63 10 22.33 3.979 78.97 39.61 20 22.42 3.962 80.79 47.02 40 22.37 3.972 81.93 44.21 3. 结 论
本研究主要聚焦于褐腐初期阶段,通过表征南方松边材内部的化学成分变化及宏观、微观结构变化等,阐明褐腐菌进入木材内部的路径及初步降解进程,得出以下结论:
(1)木材腐朽后表面颜色有偏红褐色的趋势。
(2)菌丝通过横向排列的射线薄壁细胞和轴向排列的管胞进入木材,并穿透细胞壁上的纹孔膜,从而抵达木材内部的细胞腔,并于20 d时基本完成初期定植;此时木材的质量损失速率增速最大,同时细胞壁S2层发生严重降解,细胞壁厚度损失率达到18.24%。
(3)腐朽初期,木材细胞壁中的半纤维素最先发生降解,木质素的相对含量增加。对于褐腐初期尚未发生显著降解的纤维素而言,其结晶结构发生变化;褐腐20 d时,纤维素的晶格间距最小,相对结晶度最大,可能会阻碍褐腐菌代谢产物对纤维素的分解。
-
表 1 qRT-PCR引物信息
Table 1 Information for qRT-PCR primers
基因 引物序列 产物长度/bp 18S rRNA F:TCAGCCTTGCGACCATACTCC 113 R:AACCATAAACGATGCCGACCA Actin1 F:AAGCCCAGTCCAAGAGAGGTA 107 R:ATTGTAGAAGGTGTGATGCCA Actin2 F:TAACCCCAAAGCCAACAGAGAA 140 R:CACCAGAATCCAGCACAATACC RPL40 F:AATACCTCCTGACCAACA 133 R:CTCAATAATTCCACCTCG GAPDH F:TAAAGGGTGGTGCTAAGAAG 105 R:CAATGTGAAGATCAGGAGTG UBQ F:GACTCTCCATCTGGTCCTCA 116 R:CCTTCACATTATCAATCGTATC UBQ-L10 F:AAGGCCAAGATCCAGGATAA 137 R:CGCAAGACAAGGTGAAGAGT ARF F:AGACATTACTTCCAAAACACCC 122 R:GCATCCCTTAGTTCATCCTCGT EF1α F:GCCCTACTGGACTGACCACTGA 140 R:GGATGCTACATAACCACGCTT PP2A F:GCTGCTCAGTTCAACCACACC 113 R:GCACTAAATACCGTTACCACAT RAN3 F:AAGAACAGGCAGGTGAAGGCA 120 R:ACGGGCAAGGTAGAGAAAAGG RPL23 F:GAGCCAAGAACCTTTACATC 100 R:TTGACAGTTGCCATCACCAT TUA F:AGTCTACCCATCCCCACAAG 104 R:CAAGGAGAACTGCCACATCA ACS F:TCGGAGCCTGGATGGTTTAG 135 R:TTGCCAACTTTGTTTCTTCCTT ACO F:GTTCCTCCTATGCGTCATTCCA 118 R:TCCTATTACCATCGGTTTGAGC 表 2 内参基因PCR引物序列及相关信息
Table 2 PCR primer sequences and related information for internal reference genes
基因 扩增效率(E) 决定系数(R2) 18S rRNA 1.811 0.999 29 ACT1 1.978 0.999 39 ACT2 1.965 0.999 59 RPL40 1.903 0.999 21 GAPDH 1.954 0.999 27 UBQ 1.926 0.999 25 UBQ-L10 2.037 0.999 14 ARF 1.958 0.999 21 EF1α 1.989 0.999 37 PP2A 1.981 0.999 26 RAN3 1.994 0.999 27 RPL23 1.931 0.999 25 TUA 1.924 0.999 55 表 3 13个内参基因表达稳定性的NormFinder分析结果
Table 3 NormFinder analysis result of expression stability of 13 internal reference genes
基因 切花开放至衰老不同时期 盛开期不同组织 稳定值(M) 排名 M 排名 18S rRNA 1.204 12 1.311 12 ACT1 0.357 4 0.328 4 ACT2 0.743 10 0.236 2 RPL40 0.959 11 0.862 8 GAPDH 0.454 6 0.978 9 UBQ 0.481 7 0.116 1 UBQ-L10 1.699 13 1.903 13 ARF 0.222 3 0.333 5 EF1α 0.391 5 0.760 7 PP2A 0.173 2 0.509 6 RAN3 0.160 1 0.274 3 RPL23 0.504 8 1.205 10 TUA 0.665 9 1.240 11 表 4 13个内参基因在‘桃花飞雪’切花开放至衰老不同时期花瓣中的表达稳定性综合排名
Table 4 Overall ranking of expression stability values of 13 internal reference genes in petals at different times from opening to senescence
基因 ΔCt Bestkeeper NormFinder geNorm RefFinder 18S rRNA 12 1 12 11 9 ACT1 7 6 4 8 7 ACT2 10 11 10 10 11 RPL40 11 12 11 12 12 GAPDH 4 4 6 1 2 UBQ 6 3 7 3 4 UBQ-L10 13 13 13 13 13 ARF 2 9 3 5 3 EF1α 5 5 5 6 6 PP2A 3 10 2 7 5 RAN3 1 8 1 1 1 RPL23 8 7 8 4 10 TUA 9 8 9 9 8 注:表中数据为排名次序。下同。 表 5 13个内参基因在‘桃花飞雪’不同组织中的表达稳定性综合排名
Table 5 Overall ranking of expression stability values of 13 internal reference genes in different tissues
基因 ΔCt Bestkeeper NormFinder geNorm RefFinder 18S rRNA 12 11 12 12 12 ACT1 5 2 4 5 5 ACT2 4 3 2 6 4 RPL40 8 12 8 7 9 GAPDH 9 1 9 9 7 UBQ 1 4 1 4 1 UBQ-L10 13 13 13 13 13 ARF 3 7 5 1 2 EF1α 7 8 7 8 8 PP2A 6 9 6 1 6 RAN3 2 6 3 3 3 RPL23 10 10 10 10 11 TUA 11 5 11 11 10 -
[1] 李嘉珏. 中国牡丹与芍药[M]. 北京: 中国林业出版社, 1999. Li J J. Chinese tree peony and herbaceous peony[M]. Beijing: China Forestry Publishing House, 1999.
[2] 于晓南. 观赏芍药[M]. 北京: 中国林业出版社, 2019. Yu X N. Ornamental herbaceous peony[M]. Beijing: China Forestry Publishing House, 2019.
[3] 王哲, 史国安, 马雪情, 等. 芍药‘桃花飞雪’开花衰老期间乙烯代谢生理机制的研究[J]. 园艺学报, 2014, 41(11): 2268−2274. Wang Z, Shi G A, Ma X Q, et al. The ethylene metabolism in flowers of Chinese peony ‘Taohuafeixue’ during opening and senescence[J]. Acta Horticulturae Sinica, 2014, 41(11): 2268−2274.
[4] Lu J Y, Zhang G F, Ma C, et al. The F-box protein RhSAF destabilizes the gibberellic acid receptor RhGID1 to mediate ethylene-induced petal senescence in rose[J]. Plant Cell, 2024, 36(5): 1736−1754. doi: 10.1093/plcell/koae035
[5] Wang T, Sun Z, Wang S Q, et al. DcWRKY33 promotes petal senescence in carnation (Dianthus caryophyllus L.) by activating genes involved in the biosynthesis of ethylene, abscisic acid and accumulation of reactive oxygen species[J]. The Plant Journal: for Cell and Molecular Biology, 2023, 113(4): 698−715. doi: 10.1111/tpj.16075
[6] Wang M, Wang M, Ni C, et al. Differences in ethylene sensitivity, expression of ethylene biosynthetic genes and vase life among carnation varieties[J]. Ornamental Plant Research, 2024, 4: e004.
[7] Khan S, Alvi A F, Saify S, et al. The ethylene biosynthetic enzymes, 1-aminocyclopropane-1-carboxylate (ACC) synthase (ACS) and ACC oxidase (ACO): the less explored players in abiotic stress tolerance[J]. Biomolecules, 2024, 14(1): 90. doi: 10.3390/biom14010090
[8] 李芳. 芍药花乙烯代谢及信号转导主要基因的克隆与分析[D]. 洛阳: 河南科技大学, 2013. Li F. Cloning and analysis of genes encoding ethylene biosynthesis and signal transduction elements from herbaceous peony (Paeonia lactiflora Pall.)[D]. Luoyang: Henan University of Science and Technology, 2013.
[9] 肖士奎, 李芳, 张文婷, 等. 芍药ACS基因的克隆及其蛋白质原核表达[J]. 华北农学报, 2022, 37(5): 36−44. Xiao S K, Li F, Zhang W T, et al. Cloning and prokaryotic expression of ACS gene from Paeonia lactiflora Pall. [J]. Acta Agriculturae Boreali-Sinica, 2022, 37(5): 36−44.
[10] Ashraf J, Zuo D, Wang Q, et al. Recent insights into cotton functional genomics: progress and future perspectives[J]. Plant Biotechnology Journal, 2018, 16(3): 699−713. doi: 10.1111/pbi.12856
[11] Wagner E M. Monitoring gene expression: quantitative real-time RT-PCR[J]. Methods in Molecular Biology, 2013, 1027: 19−45.
[12] Huggett J, Dheda K, Bustin S, et al. Real-time RT-PCR normalization; strategies and considerations[J]. Genes and Immunity, 2005, 6(4): 279−284. doi: 10.1038/sj.gene.6364190
[13] Peng S J, Ali S I, Hu X L, et al. Advancements in reference gene selection for fruit trees: a comprehensive review[J]. International Journal of Molecular Sciences, 2024, 25(2): 1142. doi: 10.3390/ijms25021142
[14] 李健. 芍药实时定量PCR内参基因的筛选和验证[J]. 分子植物育种, 2017, 15(7): 2544−2549. Li J. Selection and validation of reference genes for quantitative real-time PCR in herbaceous peony[J]. Molecular Plant Breeding, 2017, 15(7): 2544−2549.
[15] 梁蕤, 徐雷锋, 毕蒙蒙, 等. 柠檬色百合实时荧光定量PCR内参基因筛选[J]. 中国农业大学学报, 2023, 28(2): 59−73. doi: 10.11841/j.issn.1007-4333.2023.02.006 Liang R, Xu L F, Bi M M, et al. Validation of reference genes for quantitative real time-PCR in Lilium leichtlinii[J]. Journal of China Agricultural University, 2023, 28(2): 59−73. doi: 10.11841/j.issn.1007-4333.2023.02.006
[16] 周琳, 张永春, 蔡友铭, 等. 彩色马蹄莲不同品种和组织qRT-PCR内参基因筛选[J]. 分子植物育种, 2020, 18(12): 3971−3979. Zhou L, Zhang Y C, Cai Y M, et al. Screening of qRT-PCR reference genes in different varieties and tissues of Zantedeschia hybrida[J]. Molecular Plant Breeding, 2020, 18(12): 3971−3979.
[17] 李雪, 潘学军, 张文娥, 等. 核桃内参基因实时荧光定量PCR表达稳定性评价[J]. 植物生理学报, 2017, 53(9): 1795−1802. Li X, Pan X J, Zhang W E, et al. Stability evaluation of reference genes for quantitative real-time PCR analysis in walnut (Juglans spp.)[J]. Plant Physiology Journal, 2017, 53(9): 1795−1802.
[18] 王荣花, 刘雅莉, 李嘉瑞. 不同发育阶段牡丹和芍药切花开花生理特性的研究[J]. 园艺学报, 2005, 32(5): 861. doi: 10.3321/j.issn:0513-353X.2005.05.019 Wang R H, Liu Y L, Li J R. Studies on the blossom physiology in the different development stage of peony and Chinese peony flower[J]. Acta Horticulturae Sinica, 2005, 32(5): 861. doi: 10.3321/j.issn:0513-353X.2005.05.019
[19] Gasic K, Hernandez A, Korban S S. RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction[J]. Plant Molecular Biology Reporter, 2004, 22(4): 437−438. doi: 10.1007/BF02772687
[20] 张玉芳, 赵丽娟, 曾幼玲. 基因表达研究中内参基因的选择与应用[J]. 植物生理学报, 2014, 50(8): 1119−1125. Zhang Y F, Zhao L J, Zeng Y L. Selection and application of reference genes for gene expression studies[J]. Plant Physiology Journal, 2014, 50(8): 1119−1125.
[21] 王彦杰, 董丽, 张超, 等. 牡丹实时定量PCR分析中内参基因的选择[J]. 农业生物技术学报, 2012, 20(5): 521−528. Wang Y J, Dong L, Zhang C, et al. Reference gene selection for real-time quantitative PCR normalization in tree peony (Paeonia suffruticosa Andr.)[J]. Journal of Agricultural Biotechnology, 2012, 20(5): 521−528.
[22] Untergasser A, Ruijter J M, Benes V, et al. Web-based LinRegPCR: application for the visualization and analysis of (RT)-qPCR amplification and melting data[J]. BMC Bioinformatics, 2021, 22(1): 398.
[23] Ramakers C, Ruijter J M, Deprez R H L, et al. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data[J]. Neuroscience Letters, 2003, 339(1): 62−66. doi: 10.1016/S0304-3940(02)01423-4
[24] 王雨, 裴行杰, 傅钇钧, 等. 基于全基因组鉴定华石斛优良内参基因[J]. 热带作物学报, 2023, 44(7): 1365−1372. Wang Y, Pei X J, Fu Y J, et al. Genome-wide identification of superior reference genes in Dendrobium sinense[J]. Chinese Journal of Tropical Crops, 2023, 44(7): 1365−1372.
[25] Vandesompele J, de Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes[J]. Genome Biology, 2002, 3(7): 341.
[26] Andersen C L, Ledet-Jensen J, Ørntoft T F. Normalization of real-time quantitative RT-PCR data: a model based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets[J]. Cancer Research, 2004, 64(15): 5245−5250. doi: 10.1158/0008-5472.CAN-04-0496
[27] Pfaffl M W, Tichopád A, Prgomet C, et al. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations[J]. Biotechnology Letters, 2004, 26(6): 509−515. doi: 10.1023/B:BILE.0000019559.84305.47
[28] Xie F, Wang J, Zhang B. RefFinder: a web-based tool for comprehensively analyzing and identifying reference genes[J]. Functional & Integrative Genomics, 2023, 239(2): 125.
[29] Shabanian S, Esfahani N M, Karamian R, et al. Physiological and biochemical modifications by postharvest treatment with sodium nitroprusside extend vase life of cut flowers of two gerbera cultivars[J]. Postharvest Biology and Technology, 2018, 137: 1−8. doi: 10.1016/j.postharvbio.2017.11.009
[30] 李文阳, 马梦迪, 郭红卫. 植物激素乙烯作用机制的最新进展[J]. 中国科学: 生命科学, 2013, 43(10): 854−863. doi: 10.1360/052013-284 Li W Y, Ma M D, Guo H W. Advances in the action of plant hormone ethylene[J]. Scientia Sinica Vitae, 2013, 43(10): 854−863. doi: 10.1360/052013-284
[31] Stéphanie G, Mélanie M, Jérôme P, et al. Normalization of qRT-PCR data: the necessity of adopting a systematic, experimental conditions-specific, validation of references[J]. Journal of Experimental Botany, 2009, 60(2): 487−493. doi: 10.1093/jxb/ern305
[32] Zhang L, He L L, Fu Q T, et al. Selection of reliable reference genes for gene expression studies in the biofuel plant Jatropha curcas using real-time quantitative PCR[J]. International Journal of Molecular Sciences, 2013, 14(12): 24338−24354. doi: 10.3390/ijms141224338
[33] Luciana D, Barry C, Julia W, et al. FORMOSA controls cell division and expansion during floral development in Antirrhinum majus[J]. Planta, 2009, 229(6): 1219−1229. doi: 10.1007/s00425-009-0910-x
[34] Li C Q, Hu L Z, Wang X Q, et al. Selection of reliable reference genes for gene expression analysis in seeds at different developmental stages and across various tissues in Paeonia ostii.[J]. Molecular Biology Reports, 2019, 46(6): 6003−6011. doi: 10.1007/s11033-019-05036-7
[35] Li M, Xing Q, Li H G, et al. Selection of reference genes for quantitative real-time PCR analysis in cucumber (Cucumis sativus L.), pumpkin (Cucurbita moschata Duch.) and cucumber-pumpkin grafted plants[J]. Peer Journal, 2019, 7: e6536. doi: 10.7717/peerj.6536
[36] 尚申申, 樊琳婷, 周爽, 等. 伊藤牡丹‘巴茨拉’内参基因筛选及花瓣衰老相关基因的表达分析[J]. 植物生理学报, 2023, 59(1): 153−164. Shang S S, Fan L T, Zhou S, et al. Screening of reference genes and expression analysis of petal senescence associated genes in Itoh peony ‘Bartzella’ cut flowers[J]. Plant Physiology Journal, 2023, 59(1): 153−164.
[37] Depaepe T, van der Straeten D. Tools of the ethylene trade: a chemical kit to influence ethylene responses in plants and its use in agriculture[J]. Small Methods, 2020, 4: 1900267. doi: 10.1002/smtd.201900267
[38] Yan X Y, Zhao J H, Deng M H, et al. Two key enzyme genes associated with ethylene bio-synthesis, LbACS and LbACO, are involved in the regulation of lily flower senescence[J]. Scientia Horticulturae, 2024, 330: 113045. doi: 10.1016/j.scienta.2024.113045
[39] Pattyn J, John V H, van de Poel B. The regulation of ethylene biosynthesis: a complex multilevel control circuitry[J]. New Phytologist, 2021, 229: 770−782. doi: 10.1111/nph.16873
[40] Ryo N, Niki T, Ichimura K. Differential regulation of two 1-aminocyclopropane-1-carboxylate oxidase (ACO) genes, including the additionally cloned DcACO2, during senescence in carnation flowers[J]. Postharvest Biology and Technology, 2022, 183: 111752. doi: 10.1016/j.postharvbio.2021.111752
[41] Yokotani N, Nakano R, Imanishi S, et al. Ripening-associated ethylene biosynthesis in tomato fruit is autocatalytically and developmentally regulated[J]. Journal of Experimental Botany, 2009, 60(12): 3433−3442. doi: 10.1093/jxb/erp185
-
期刊类型引用(2)
1. 李建建,贺宸靖,黄小平,向太和. 植物长链非编码RNA调控发育与胁迫应答的研究进展. 生物技术通报. 2023(01): 48-58 .  百度学术
百度学术
2. 黄杰,陈勇,高日芳,毛莹莹,郑柏艳,张帆涛,谢建坤. 长链非编码RNA:与植物发育和胁迫响应相关的新型调控因子. 江西师范大学学报(自然科学版). 2023(06): 615-625 .  百度学术
百度学术
其他类型引用(3)








 下载:
下载: