Cloning and expression characteristics analysis of BpTCP2 gene in Betula platyphylla
-
摘要:目的通过克隆白桦BpTCP2基因,分析其序列特征,并检测该基因在不同组织部位、激素处理及非生物胁迫下的基因表达特性,为揭示BpTCP2在植物生长发育及逆境应答中的作用提供理论依据。方法通过PCR技术克隆BpTCP2基因的全长,并对得到的cDNA序列进行生物信息学分析;采用实时荧光定量PCR技术分析该基因的组织表达特性,以及在外源植物激素(ABA、IAA、BR、JA、SA)处理和非生物胁迫(CdCl2、NaCl、NaHCO3、PEG)下的响应模式。结果成功克隆了BpTCP2基因的cDNA序列;生物信息学分析发现BpTCP2含有高度保守的bHLH结构域,属于PCF亚类。qRT-PCR结果显示,BpTCP2基因在木质部、幼嫩的叶片和茎节中表达量较高;在CdCl2、NaCl、NaHCO3处理下,该基因持续上调表达;且5种外源激素均能诱导该基因表达。结论BpTCP2基因参与白桦木质部的形成,影响叶片及初期茎节的发育,并能在各激素、重金属、盐、碱的诱导下表达。Abstract:ObjectiveThe BpTCP2 gene was cloned, sequence characteristics were analyzed, and the gene expression characteristics of the gene under different tissues, hormone treatments and abiotic stress were detected, which provided a reference for revealing the function of BpTCP2 in plant growth and development and stress response.MethodThe full-length cDNA sequence of BpTCP2 gene was cloned by PCR technology and its cDNA sequence characteristics were analyzed by bioinformatics. Real-time quantitative PCR was used to analyze expression level of the gene in different tissues and the responses of exogenous phytohormones (ABA, IAA, BR, JA, SA) and abiotic stress (CdCl2, NaCl, NaHCO3, PEG).ResultThe full-length cDNA sequence of BpTCP2 gene was successfully cloned. Bioinformatics analysis found that BpTCP2 contained a highly conserved bHLH domain and BpTCP2 belongs to TCP-P subclass; qRT-PCR analysis showed that the expression of BpTCP2 gene was higher in xylem, young leaves and stem segments. Under the treatment of CdCl2, NaCl and NaHCO3, the gene was up-regulated. Five exogenous phytohormones could induce the expression of the gene.ConclusionThe BpTCP2 gene was involved in the development of stems of Betula platyphylla, and affects the development of leaves and early stem segments and responds to various hormones and heavy metals, salts and alkalis.
-
Keywords:
- Betula platyphylla /
- BpTCP2 /
- bioinformatics /
- gene cloning /
- expression characteristics
-
转录因子通过调节动物和植物基因的时空表达水平,来调控生物体的细胞增殖、生长发育及免疫反应过程[1]。TCP基因家族编码植物特有的转录调控因子,该家族成员在植物生长[2]、花瓣的不对称性[3-4]、细胞分裂[5]、叶形态发生[6-7]、衰老[8-9]、胚胎生长[10]、昼夜节律[2]中起着关键作用。基于蛋白质的DNA结构域不同,该家族成员分为Class I(PCF类)和Class II(CIN和CYC/TB1)两类。目前,Class I类基因功能在草本植物中研究的较为深入。在拟南芥(Arabidopsis thaliana)中,AtTCP11通过上调VND7基因的表达,引起维管发育缺陷[11];AtTCP14通过细胞增殖控制叶的形态发生;AtTCP20通过抑制茉莉酸(JA)在植物体内的合成,降低其抑制细胞增殖的能力,从而使细胞大小发生改变[12],同时转AtTCP20::EAR拟南芥表现出发芽延迟、茎尖和根尖分生组织生长速度放缓、子叶变黄,下胚轴区域膨胀等特征[13]。
TCP基因参与多种激素信号传递过程,调控植物形态发育。在拟南芥中,AtTCP5、AtTCP13和AtTCP17通过PIFs依赖途径和不依赖PIFs途径两种方式来促进生长素的合成,进而介导拟南芥下胚轴在遮荫条件下的伸长[14];AtTCP3和AtTCP15通过调控生长素响应相关基因SHY2/IAA3和SAUR的表达参与生长素信号的转导[15-16];AtTCP9通过改变JA的代谢从而改变根长[17];在陆地棉(Gossypium hirsutum)中,GhTCP1基因通过促进JA的合成而使棉花纤维伸长并促进根毛发育[18]。AtTCP14和AtTCP15参与细胞分裂素(CK)信号途径的应答,影响细胞分裂[19];I类TCP转录因子家族参与赤霉素(GA)信号转导,研究发现GA通过刺激DELLA蛋白的降解来控制茎和节间伸长,而DELLA蛋白直接调节植物特异性I类TCP转录因子家族的活性,从而控制细胞增殖,同时拟南芥tcp8、tcp14、tcp15、tcp22这4个基因的突变体表现出严重的侏儒症和对GA作用的反应敏感性降低的性状[20]。目前对PCF亚类基因的研究主要集中在草本植物上,而在木本植物中研究较少。
白桦(Betula platyphylla),桦木科(Betulaceae)落叶类乔木,白桦树干修直、洁白,且耐严寒,生长较快,是我国东北地区珍贵阔叶树种之一。本研究以白桦为试材,克隆了BpTCP2基因编码区的全长序列,对其进行生物信息学分析,同时采用qRT-PCR分析该基因在白桦不同组织部位的表达特征以及对植物激素处理和非生物胁迫的应答机制,为揭示白桦的生长发育、抗逆机制,以及培育优良的白桦新品系提供参考。
1. 材料与方法
1.1 材 料
1.1.1 植物材料
材料于2018年夏季取自东北林业大学林木遗传育种白桦强化育种基地栽植的2年生白桦组培苗木,依次选取5株的顶芽、腋芽、第1到第7茎节(幼嫩到成熟)、成熟茎节的木质部和韧皮部、第1到第13片叶(包括叶片从幼嫩到衰老过程)。以上植物材料经液氮速冻后,放入− 80 ℃超低温冰箱保存,用于RNA的提取和研究BpTCP2在白桦不同组织部位的表达。
从白桦强化育种基地采集的白桦全同胞家系种子通过水培萌发,待长出两片子叶时,分别取10株长势均一的植株,放置在涂有相应外源激素和非生物胁迫试剂的WPM培养基中。激素处理的种类及浓度分别为:50 mg/L IAA,100 μmol/L ABA,0.2 mg/L油菜素类固醇(BR),1 μmol/L JA,350 μmol/L 水杨酸(SA)。非生物胁迫处理的浓度分别为:0.4 mol/L NaCl,0.3 mol/L NaHCO3,150 μmol/L CdCl2,20% PEG。分别在0、2、4、6、12和24 h取材并放置于1.5 mL离心管中,液氮速冻后,放入− 80 ℃超低温冰箱内保存,用于RNA提取。
1.1.2 试 剂
RNA提取试剂盒为通用植物总RNA提取试剂盒(离心柱型),购置于北京百泰克生物技术有限公司,反转录试剂盒为ReverTra Ace® qPCR RT Master Mix with gDNA Remover试剂盒,购置于东洋纺(上海)生物科技有限公司,TransStart Top Green qPCR SuperMix(Perfect Real Time染料法实时荧光定量)购置全式金生物(北京)有限公司,Topo试剂盒为pENTRTM/D-TOPO® Cloning Kit购置于invitrogen公司,大肠杆菌Trans1-T1感受态细胞购自世国生物科技(哈尔滨)有限公司。引物由擎科(哈尔滨)公司合成。
1.2 方 法
1.2.1 白桦BpTCP2基因的克隆
根据白桦基因组和转录组序列比对获得BpTCP2基因的序列设计引物,上、下游引物序列分别为5′-CACCGACATGGCAGAGAGCAAGC-3′、5′-ATTCTTCTACTGCCTTGACC-3′。以白桦不同组织部位的cDNA为模板,PCR扩增BpTCP2基因目标片段,反应体系如下:10 × KOD Buffer 1.7 μL,2 mmol/L dNTPs 1.7 μL,MgSO4 0.8 μL,cDNA模板0.4 μL,上、下游引物各为10 mmol/L 0.6 μL,KOD Plus 0.4 μL,ddH2O补足20 μL。PCR扩增程序为94 ℃预变性2 min;94 ℃变性45 s,58 ℃退火45 s,68 ℃延伸2 min,35个循环;68 ℃延伸10 min。PCR产物经1.0% 琼脂糖凝胶电泳检测,并进行胶回收纯化,将纯化产物进行Topo反应,连接体系为:胶回收产物1 μL,Topo Vector 0.5 μL,Salt Solution 0.5 μL,用ddH2O补足反应体系至3 μL。25 ℃连接30 min,连接反应完成后,通过热击法将连接产物转化到大肠杆菌Trans1-T1感受态细胞中。对获得的单克隆进行PCR检测,条带位置正确后送公司测序,保存经测序对比正确的菌液。
1.2.2 生物信息学分析
采用NCBI ORF Finder查找序列的ORF;用在线软件BioEdit预测该基因氨基酸序列;用Blastx进行同源序列比对,搜索不同物种中的同源基因及蛋白,并先将氨基酸序列利用Clustal X进行多序列比对的分析,借助MEGA 5.1软件的Neighbor-Joining算法,1 000次重复,以默认参数构建系统发育进化树[21]。
1.2.3 Real-time PCR分析BpTCP2基因的表达特性
使用RNA提取试剂盒提取白桦不同组织部位(顶芽、腋芽、木质部、韧皮部、1 ~ 7茎节、1 ~ 13叶片)、5种外源植物激素(IAA、ABA、BR、JA、SA)及4种非生物胁迫(NaCl、NaHCO3、CdCl2、PEG)处理后的整株材料的总RNA,并进行反转录。根据克隆获得的BpTCP2基因的全长cDNA序列设计定量引物,同时选用α-Tubulin作为内参基因[21](引物见表1)。获得的cDNA稀释10倍作为qPCR模板。实时定量PCR反应体系为:6 μL Top qMix,0.24 μL Passive Reference Dye,2 μL cDNA,10 μmol/L的上、下游引物各0.24 μL,用ddH2O补足反应体系至12 μL。扩增反应在ABI PRISM® 7500荧光定量PCR仪上完成,反应程序为94 ℃预变性30 s,94 ℃变性5 s,56 ℃退火15 s,72 ℃延伸34 s(45个循环),绘制溶解曲线的温度为95 ℃持续15 s,60 ℃持续1 min,95 ℃持续30 s。所有样品均进行3次重复,采用– ΔΔCt方法进行基因的相对定量分析。
表 1 实时荧光定量PCR引物序列Table 1. Real-time PCR primer sequences引物名称 Primer name 上游引物序列(5′→3′) Forward primer (5′→3′) 下游引物序列(5′→3′) Reverse primer (5′→3′) BpTCP2 5′-GCTTGCATACAAAGATGGAAGG-3′ 5′-GGAAAAGCTCAATGGACCCAG-3′ α-Tubulin 5′-GCACTGGCCTCCAAGGAT-3′ 5′-TGGGTCGCTCAATGTCAAGG-3′ 2. 结果与分析
2.1 BpTCP2基因的克隆
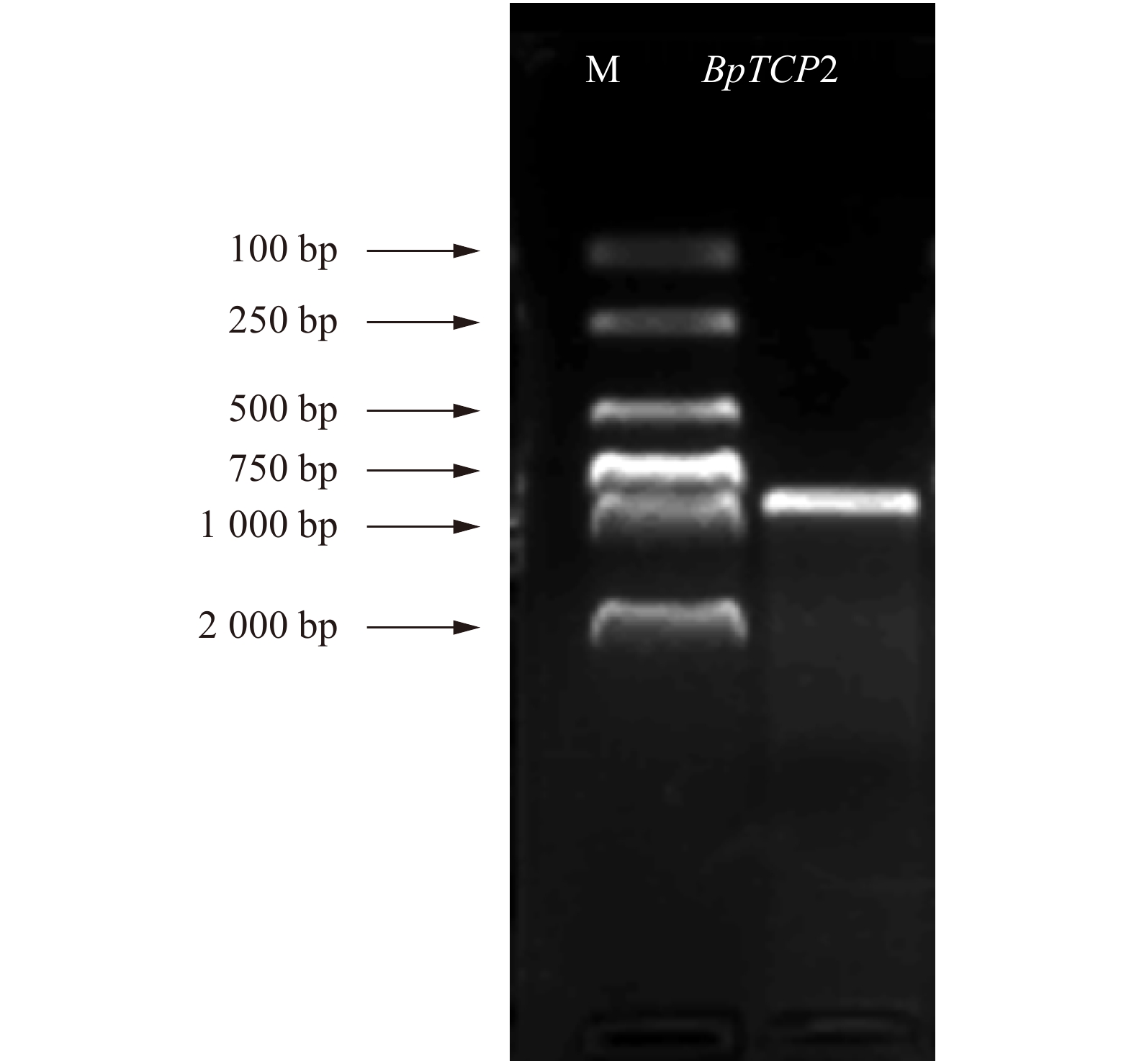
以白桦不同组织部位的cDNA为模板,根据白桦基因组序列和转录组数据比对获得BpTCP2基因序列设计特异性引物,PCR扩增其全长cDNA序列,扩增产物于1.0%琼脂糖凝胶电泳检测(图1),纯化后的胶回收产物通过Topo反应进行菌液PCR检测,结果显示在813 bp处获得特异性条带,与预期目标条带大小一致(图2)。
2.2 白桦BpTCP2基因的生物信息学分析
2.2.1 白桦BpTCP2基因编码区序列分析
利用BLASTx及NCBI ORF Finder预测BpTCP2基因的ORF全长,利用生物学软件BioEdit预测BpTCP2基因的ORF全长为804 bp,编码267个氨基酸(图3)。
2.2.2 白桦BpTCP2基因结构域分析
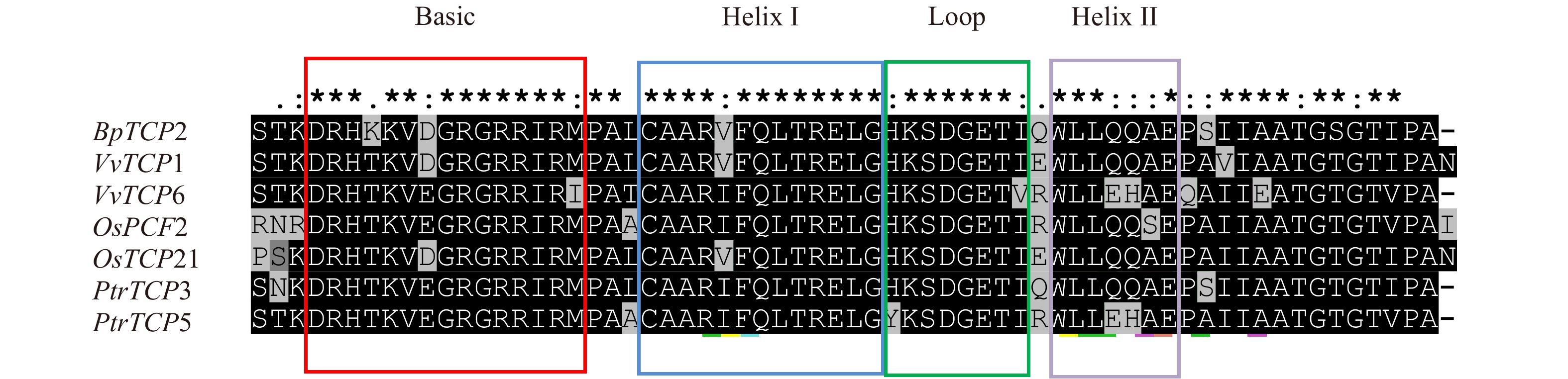
分别选取拟南芥、葡萄(Vitis vinifera)、水稻(Oryza sativa)、毛果杨(Populus trichocarpa)4个物种PCF亚类基因各2条与BpTCP2基因进行氨基酸的多序列比对分析,结果如图4所示,BpTCP2基因与其他物种PCF亚类基因的氨基酸序列均含有TCP保守结构域bHLH[22]。bHLH基序由44个氨基酸组成,包括一个能与DNA结合的碱性区域和α螺旋1-环-α螺旋2(Helix1- Loop-Helix2)组成[23-24],其中Basic结构域含有16个氨基酸,helixI、loop和helixII结构域分别含有11、8和9个氨基酸。从比对结果可以看出无论是Basic结构域还是其他3个结构域均高度保守。
2.2.3 白桦BpTCP2基因的进化树分析
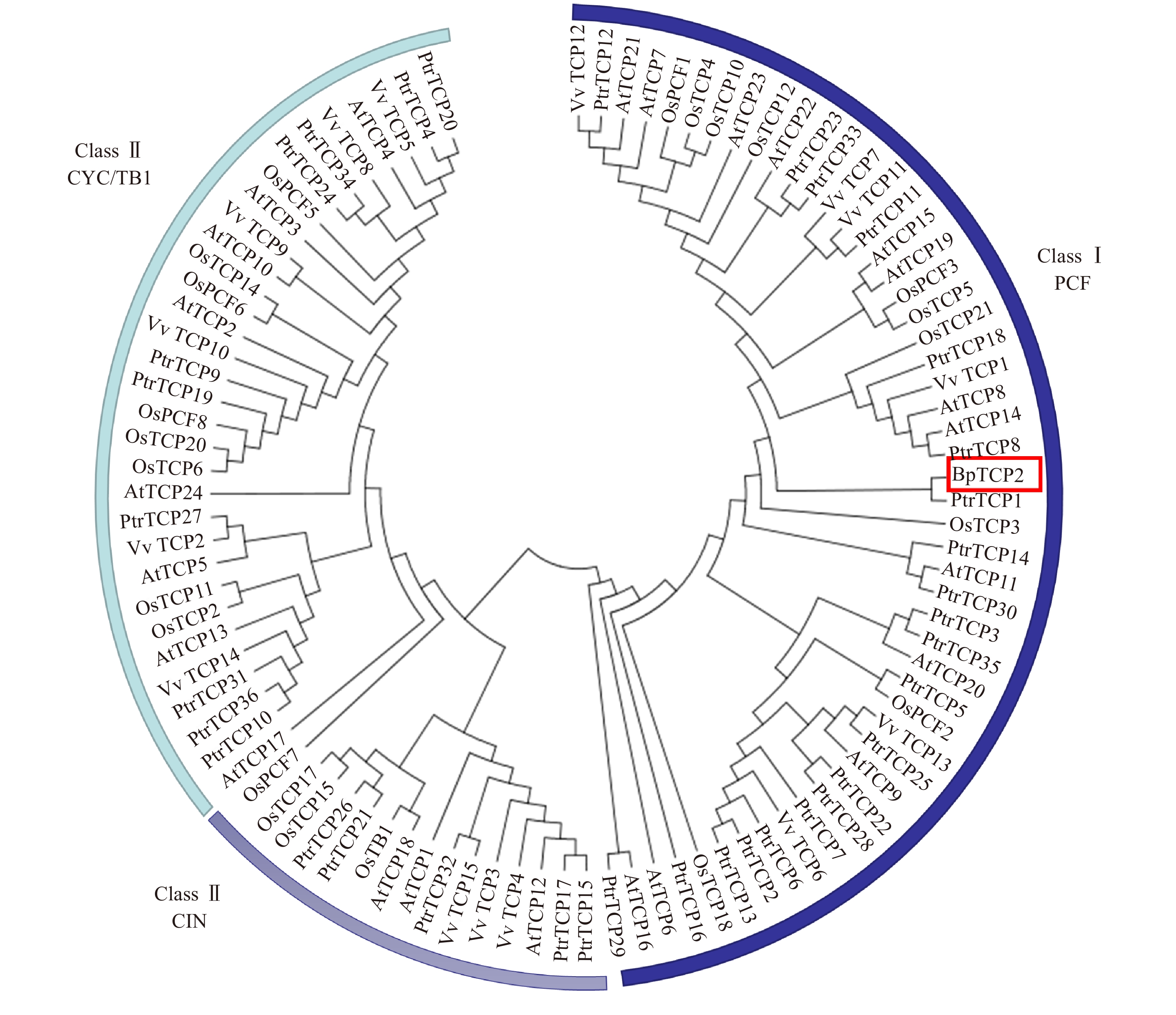
利用MEGA 5.1对白桦BpTCP2与拟南芥、水稻、葡萄及毛果杨TCP的蛋白序列进行聚类分析,结果如图5所示,97个TCP成员被清楚的分为Class I(PCF)和Class II(CIN和CYC/TB1)两大类,其中BpTCP2属于PCF亚类,并与毛果杨的TCP1基因相似性较高。
2.3 BpTCP2基因的组织表达特性分析
2.3.1 不同组织部位
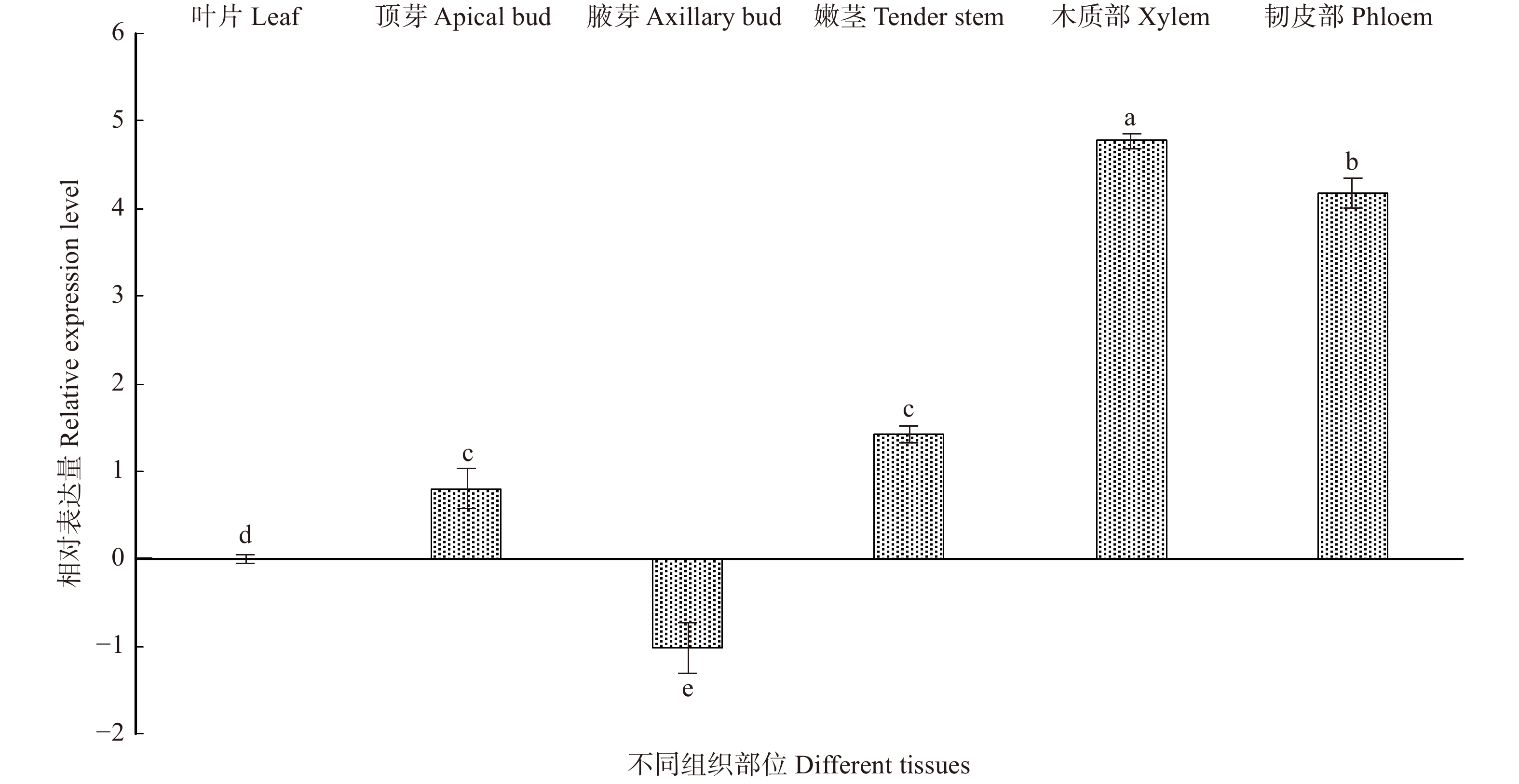
以叶为对照,分析了BpTCP2基因在顶芽、腋芽、嫩茎、木质部及韧皮部的表达情况(图6),结果表明:该基因在腋芽中表达量最低,而在其他4个组织部位中均上调表达,尤其在木质部和韧皮部中,表达量分别上调32倍和16倍。
2.3.2 不同叶发育阶段
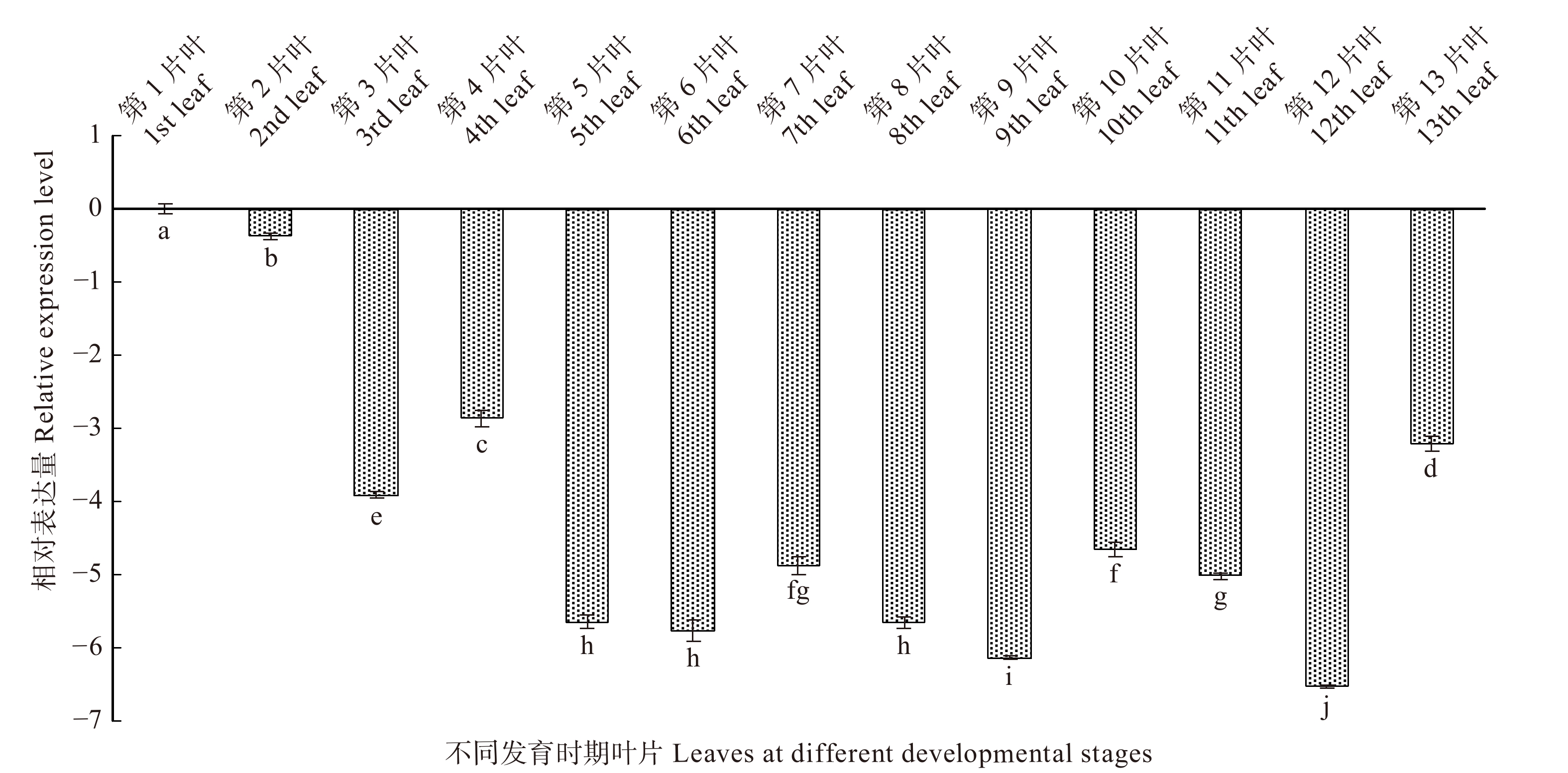
根据白桦叶片的发育情况,分别选取从幼嫩到衰老整个发育阶段的叶片(图7),并以第1片叶为对照,分析了BpTCP2基因在叶片不同发育时期的表达情况(图8),结果表明:该基因随着叶龄的增加整体呈现下调的趋势;在前期发育过程中(第1 ~ 4片叶)BpTCP2表达量呈现先下调后上调的趋势;在中期发育过程(第5 ~ 9片叶),该基因表达量变化并不明显(< 2倍),但低于对照;在后期发育过程(第10 ~ 13片叶)中,该基因持续下调,在第12片叶时下调达到峰值,为对照的128倍,在第13片叶时表达量略有回升。
2.3.3 茎节发育阶段
根据白桦茎节的发育情况,选取了白桦从幼嫩到成熟的茎节如(图9),分析BpTCP2基因在茎节发育阶段的表达情况(图10),以第1茎节为对照,该基因在茎发育初期,表达量呈现持续上调的趋势,尤其是在第4茎节中表达量达到最高峰,为对照的4倍。但从第5茎节到成熟茎节其在韧皮部中的表达量变化不明显(< 2倍)。
2.4 激素应答分析
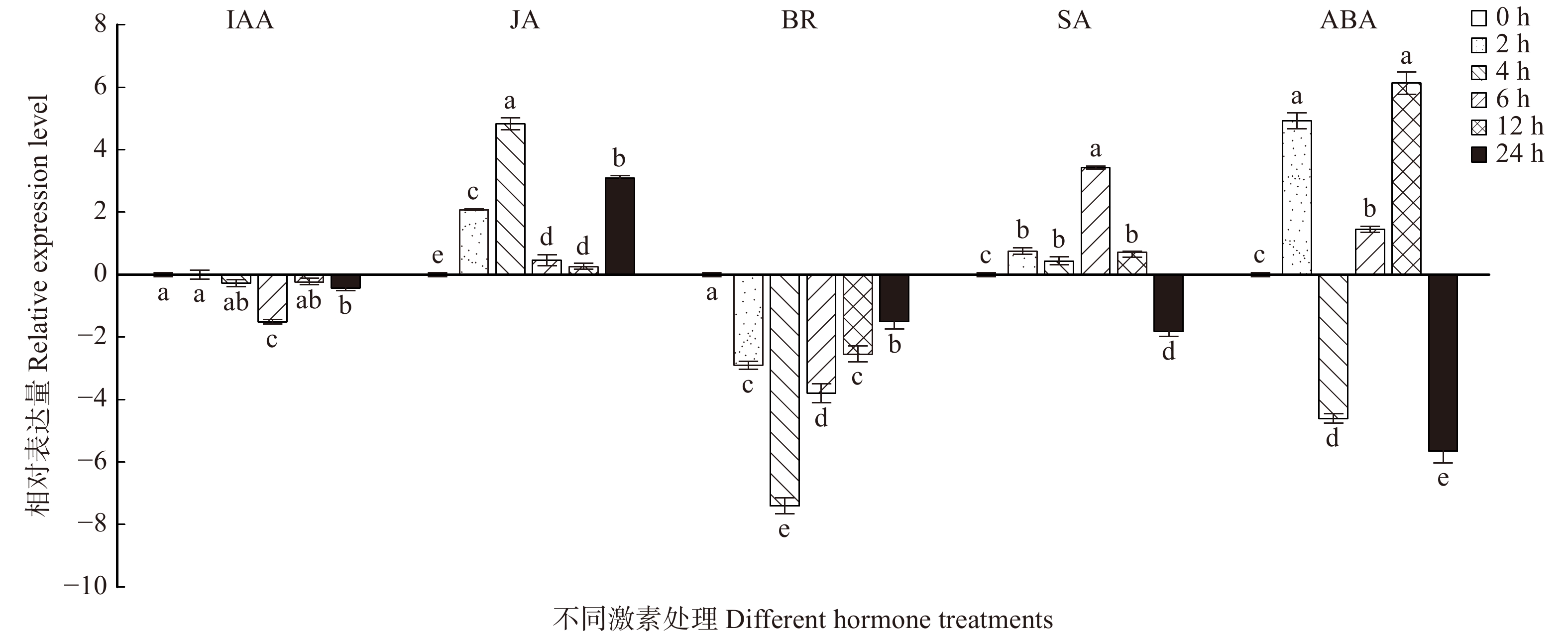
在BR、JA、SA、IAA、ABA这5种激素处理下,BpTCP2的表达情况呈现不同的表达趋势,如图11所示。在BR处理下,BpTCP2基因呈现下调表达的趋势,尤其是在4 h时表达量达到最低值,下调为对照的256倍,随着处理时间的延长,其略有上升但还是低于处理前(0 h);在JA处理过程中,除6、12 h与对照相比变化不明显外,其他时间点均呈现上调表达的趋势,尤其在4 h时,其表达量明显比对照提高了32倍;SA处理后,BpTCP2的表达量在12 h前均上调,且在6 h时达到峰值,为未处理的8倍,而在24 h时骤然下调为4倍;在IAA的处理过程中,BpTCP2基因呈现下调表达的趋势,且除了6 h外,在其他时间点BpTCP2基因的表达量没有发生明显的变化(< 2倍);在ABA处理过程中,BpTCP2的表达量呈现波动,在12 h时表达量达到最高峰,为对照的32倍,而在24 h时表达量最低,为对照的64倍。
2.5 非生物胁迫应答分析
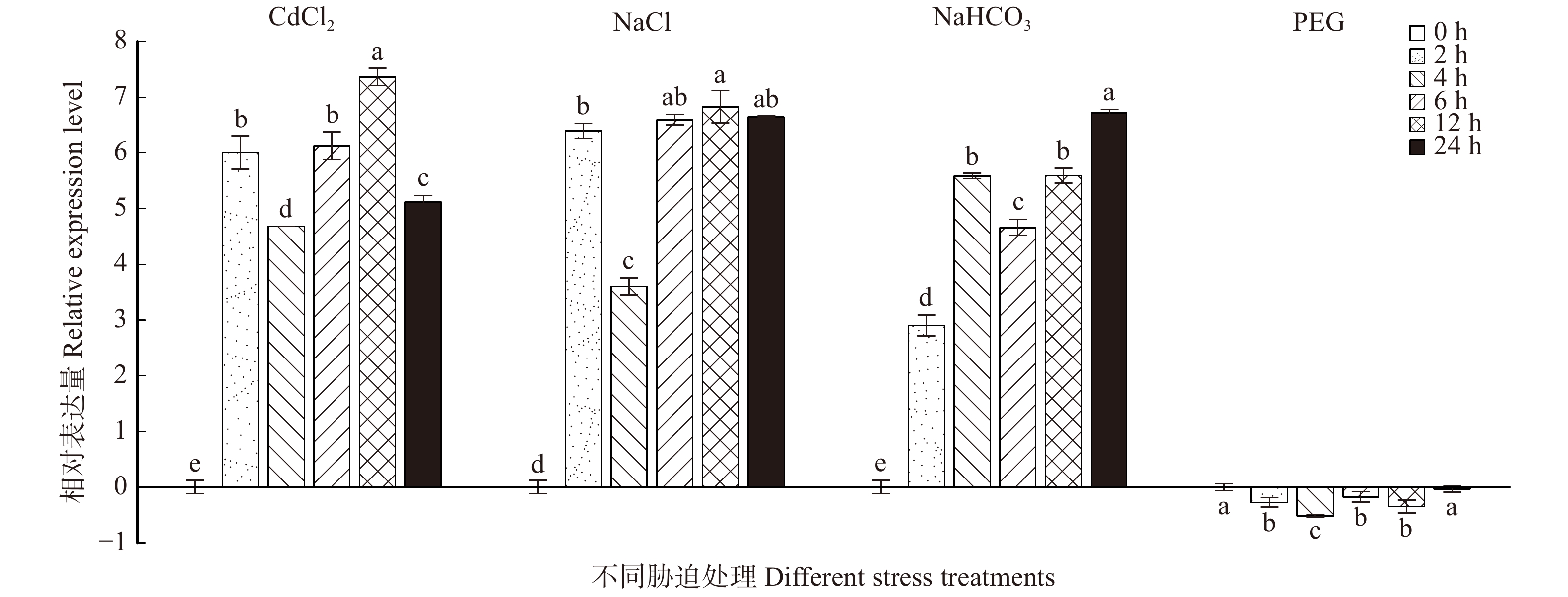
通过CdCl2、NaCl、NaHCO3、PEG 4种胁迫处理,分析BpTCP2的表达情况,结果如图12所示。其中,在CdCl2、NaCl和NaHCO3处理下,BpTCP2基因的表达模式相似且均为高表达,并且CdCl2和NaCl在处理12 h时上调表达量达到峰值,分别为对照的128倍和64倍,NaHCO3处理后表达量最高峰出现在24 h,为对照的64倍;在PEG胁迫处理过程中,BpTCP2与对照相比变化并不明显(< 2倍)。
3. 讨 论
通过对白桦的BpTCP2氨基酸序列以及葡萄、拟南芥、水稻、毛果杨TCP氨基酸序列进行系统聚类分析,根据其序列差异将其分为Class I(PCF)和Class II(CIN和CYC/TB1)两大类,其中BpTCP2属于Class I类PCF亚类。Class I类和Class II类相比,Class I类在Basic结构域上有4个氨基酸的缺失[25-26]。
分析BpTCP2在不同组织部位的特异性表达发现,该基因在嫩茎、木质部、韧皮部、顶芽中均呈上调表达,且在木质部中表达量最高,表明BpTCP2可能与木质素的生物合成相关。在安家兴等人[11]的研究中,也证明了Class I类PCF亚类的AtTCP11通过改变VND7的表达量从而影响维管束的发育。分析BpTCP2在茎节发育中的表达水平,从第1茎节到第4茎节表达量呈现持续上调的趋势,说明BpTCP2可能参与白桦茎的初期发育,Kieffer等[27]发现了AtTCP15和AtTCP14的突变体通过影响细胞增殖来影响茎节发育。在叶片发育的表达分析中,随着叶龄的增加,BpTCP2表达量整体呈现一个下调的趋势说明BpTCP2可能和延缓衰老相关,张春雷等[28]人在对拟南芥的PCF亚类基因AtTCP15、AtTCP22研究时发现,35S::TCP15-SRDX和35S::TCP22-SRDX同时表现出提前衰老的现象。
近年研究发现TCP转录因子除了在植物发育过程中发挥重要作用外,还参与植物激素及胁迫的信号传导等过程[29],同时我们对BpTCP2基因的启动子序列进行分析,也发现其含有多种激素和胁迫的应答元件(未发表),故本研究对BpTCP2基因进行了外源激素和非生物胁迫应答分析,结果表明,在植物激素处理下,BpTCP2呈现不同的表达趋势。经ABA处理后,BpTCP2呈现出先上调后下调的波动型表达模式,说明该基因参与ABA的信号应答。Mukhopadhyay等[30]对PCF亚类的OsTCP19的信号通路进行研究,发现OsTCP19通过与转录因子ABI4的互作来调控ABI3的表达从而对ABA信号通路进行调控;经BR处理后,BpTCP2整体均呈现下调表达,尤其在4 h时表达量最低,说明BpTCP2明显的响应了BR的信号且为负响应模式。DWARF4是BR生物合成中的关键酶,在拟南芥中,通过染色体免疫共沉淀发现AtTCP1可以促进DWF4的表达,使内源BR含量增加,促进植物的生长[29];在IAA处理后,BpTCP2在6 h时呈现下调表达的趋势而在其他时间点表达量变化并不明显,说明BpTCP2也响应了IAA的初期信号。在拟南芥中CIN类的AtTCP3可以引起IAA合成的负调节因子CYP83B1/SUR2的上调[31],AtTCP15(PCF类)也可间接诱导IAA3/SHY2表达[2],通过使用IAA响应原件DR5::GUS分析,AtTCP15和AtTCP3在一定程度上功能冗余,表明他们在IAA信号通路中可能发挥着相同的作用,这与BpTCP2处理后的结果类似;在JA处理后,BpTCP2除6 h、12 h与对照相比变化不明显外,其他时间点均呈现上调表达的趋势,说明BpTCP2响应了JA的信号且为正响应模式,在冯志娟等人[29]的研究中,LIPOXYGENASE2(LOX2)基因受TCP转录因子的调控且LOX2的诱导使JA积累;在SA处理下,BpTCP2的表达量在12 h前均为上调表达,但在24 h时骤然下降,说明BpTCP2响应了SA的信号,拟南芥AtTCP8(PCF亚类)与ICS1转录激活因子SARD1和WRKY28以及ICS1阻遏蛋白NAC019相互作用,ICS1基因是编码SA生物合成过程中必需的酶[32]且拟南芥中AtTCP8可以作为效应诱导免疫过程中的正向调节者[29]。
在CdCl2、NaCl、NaHCO3处理过程中,BpTCP2呈现上调表达的趋势,表明该基因在相应的非生物胁迫过程中表现出正调控模式,这一结果与水曲柳(Fraxinus mandschurica)和木薯(Manihot esculenta)的研究结果一致,水曲柳的FmTCP4和木薯的MeTCPs基因在盐胁迫下也呈现出正响应模式[33-34];在水稻中,OsTCP19的过表达导致LOX2(茉莉酸信号通路基因)的下调,从而减少水分流失和活性氧的缺失以及脂肪的积累,进而提高植株的耐盐性[30];本实验团队前期对PromTCP7::GUS拟南芥株系进行盐、旱处理,发现该基因对盐、旱途径均有不同程度响应[35]。耐盐性结果与本实验基本一致,但在PEG(干旱)处理下,BpTCP2的变化并不明显。目前,miR319的靶位点已在多个物种的TCP基因中被发现,在植物非生物胁迫应答的过程中,miR319与TCP基因的模式靶向关系为负调控[29],作为miR319的靶基因,TCP家族基因间存在功能冗余与分歧;并且在拟南芥中Viola等[36]发现,半胱氨酸(Cys-20)是AtTCP15保守结构域中的重要组成部分,AtTCP15通过其在氧化胁迫条件下发生的氧化反应从而抑制该基因的转录水平,进而参与非生物胁迫过程。
4. 结 论
本实验成功克隆了BpTCP2基因,生物信息分析表明该基因属于Class I类PCF亚类且含有高度保守的bHLH结构域;BpTCP2参与木质素的生物合成、初期茎节发育及叶片发育过程;BpTCP2参与植物外源激素(ABA、IAA、BR、JA、SA)及非生物胁迫(重金属、盐、碱)的响应。
-
表 1 实时荧光定量PCR引物序列
Table 1 Real-time PCR primer sequences
引物名称 Primer name 上游引物序列(5′→3′) Forward primer (5′→3′) 下游引物序列(5′→3′) Reverse primer (5′→3′) BpTCP2 5′-GCTTGCATACAAAGATGGAAGG-3′ 5′-GGAAAAGCTCAATGGACCCAG-3′ α-Tubulin 5′-GCACTGGCCTCCAAGGAT-3′ 5′-TGGGTCGCTCAATGTCAAGG-3′ -
[1] Ma J, Liu F, Wang Q, et al. Comprehensive analysis of TCP transcription factors and their expression during cotton (Gossypiumarboreum) fiber early development[J/OL]. Scientific Reports, 2016, 6(1): 21535 [2018−11−30]. https://doi.org/10.1038/srep21535.
[2] 冯雅岚, 熊瑛, 张均, 等. TCP转录因子在植物发育和生物胁迫响应中的作用[J]. 植物生理学报, 2018, 54(5):22−30. Feng Y L, Xiong Y, Zhang J, et al. The role of TCP transcription factors inplant development and biotic stress response[J]. Chinese Journal of Plant Physiology, 2018, 54(5): 22−30.
[3] Luo D, Carpenter R, Vincent C, et al. Origin of floral asymmetry in Antirrhinum[J]. Nature, 1996, 83: 794−799.
[4] Luo D, Carpenter R, Copsey L, et al. Control of organ asymmetry in flowers of Antirrhinum[J]. Cell, 1999, 99(4): 367−376. doi: 10.1016/S0092-8674(00)81523-8
[5] Nath U. Genetic control of surface curvature[J]. Science, 2003, 299: 1404−1407. doi: 10.1126/science.1079354
[6] Palatnik J F, Allen E, Wu X, et al. Control of leaf morphogenesis by microRNAs[J]. Nature, 2003, 425: 257−263. doi: 10.1038/nature01958
[7] Ori N, Cohen A R, Etzioni A, et al. Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato[J]. Nature Genetics, 2007, 39(6): 787−791. doi: 10.1038/ng2036
[8] Schommer C, Palatnik J F, Aggarwal P, et al. Control of Jasmonate Biosynthesis and Senescence by miR319 Targets[J/OL]. PLoS Biology, 2008, 6(9): e230 [2018−11−30]. https://doi.org/10.1371/journal.pbio.0060230.
[9] Mao C, Lu S, Lü B, et al. A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis[J]. Plant Physiology, 2017, 174(3): 1747−1763. doi: 10.1104/pp.17.00542
[10] Tatematsu K, Nakabayashi K, Kamiya Y, et al. Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana[J]. The Plant Journal, 2008, 53(1): 42−52. doi: 10.1111/tpj.2008.53.issue-1
[11] 安家兴. 拟南芥TCP11调控维管束的发育[D]. 兰州: 兰州大学, 2012. An J X. Arabidopsis thaliana TCP11 regulates the development of vascular bundles[D]. Lanzhou: Lanzhou University, 2012.
[12] Cubas P, Lauter N, Doebley J, et al. The TCP domain: a motif found in proteins regulating plantgrowth and development[J]. The Plant Journal, 1999, 18(2): 215−222. doi: 10.1046/j.1365-313X.1999.00444.x
[13] Herve C, Dabos P, Bardet C, et al. In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development[J]. Plant Physiology, 2009, 149(3): 1462−1477. doi: 10.1104/pp.108.126136
[14] 周瑜. TCP转录因子通过促进PIFs活性和生长素合成基因的表达调控避荫反应[D]. 兰州: 兰州大学, 2018. Zhou Y. TCP transcription factors regulate shade avoidance via promoting the activity of PIFs and the expression of auxin biosynthetic genes[D]. Lanzhou: Lanzhou University, 2018.
[15] Ubertimanasero N G, Lucero L E, Viola I L, et al. The class I protein AtTCP15 modulates plant development through a pathway that overlaps with the one affected by CIN-like TCP proteins[J]. Journal of Experimental Botany, 2012, 63(2): 809−823. doi: 10.1093/jxb/err305
[16] Koyama T, Mitsuda N, Seki M, et al. TCP transcription factors regulatethe activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis[J]. Plant Cell, 2010, 22(11): 3574−3588. doi: 10.1105/tpc.110.075598
[17] Danisman S, Van der Wal F, Dhondt S, et al. Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically[J]. Plant Physiology, 2012, 159(4): 1511−1523. doi: 10.1104/pp.112.200303
[18] 刘伟娜, 韩建民, 董金皋, 等. 棉花TCP家族转录因子基因GhTCP1的克隆与表达分析[J]. 棉花学报, 2010, 22(5):409−414. doi: 10.3969/j.issn.1002-7807.2010.05.005 Liu W N, Han J M, Dong J G, et al. Cloning and expression analysis of cotton TCP family transcription factor gene GhTCP1[J]. Journal of Cotton, 2010, 22(5): 409−414. doi: 10.3969/j.issn.1002-7807.2010.05.005
[19] Steiner E, Efroni I, Gopalraj M, et al. The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers[J]. Plant Cell, 2012, 24(1): 96−108. doi: 10.1105/tpc.111.093518
[20] Daviere J M, Wild M, Regnaul T, et al. Class I TCP-DELLA interactions in inflorescence shoot apex determine plant height[J]. Current Biology, 2014, 24(16): 1923−1928. doi: 10.1016/j.cub.2014.07.012
[21] 宁坤, 杨洋, 马述山, 等. 11条白桦BpSPL家族基因的生物信息学和表达分析[J]. 林业科学研究, 2016, 29(5):646−653. doi: 10.3969/j.issn.1001-1498.2016.05.004 Ning K, Yang Y, Ma S S, et al. Bioinformatics and expression analysis of 11 BpSPL family genes in Betula platyphylla[J]. Forest Research, 2016, 29(5): 646−653. doi: 10.3969/j.issn.1001-1498.2016.05.004
[22] 董京祥, 任丽, 张园, 等. 白桦BpTCPs基因家族生物信息学及时空表达分析[J]. 南京林业大学报, 2018, 42(4):113−118. Dong J X, Ren L, Zhang Y, et al. Analysis of temporal and spatial expression of BpTCPs gene family bioinformatics in Betula platensis[J]. Journal of Nanjing Forestry University, 2018, 42(4): 113−118.
[23] 王勇江, 陈克平, 姚勤. bHLH转录因子家族研究进展[J]. 遗传, 2008, 30(7):821−830. doi: 10.3321/j.issn:0253-9772.2008.07.004 Wang Y J, Chen K P, Yao Q. Research progress of bHLH transcription factor family[J]. Genetics, 2008, 30(7): 821−830. doi: 10.3321/j.issn:0253-9772.2008.07.004
[24] 刘文文, 李文学. 植物bHLH转录因子研究进展[J]. 生物技术进展, 2013, 3(1):7−11. Liu W W, Li W X. Research progress of plant bHLH transcription factors[J]. Progress in Biotechnology, 2013, 3(1): 7−11.
[25] Manassero N G, Viola I L, Welchen E, et al. TCP transcription factors: architectures of plant form[J]. Biomolecular Concepts, 2013, 4(2): 111−127.
[26] De Paolo S, Gaudio L, Aceto S. Analysis of the TCP genes expressed in the inflorescence of the orchid Orchis italica[J/OL]. Scientific Reports, 2015, 5(1): 16265[2018−11−30]. https://doi.org/10.1038/srep16265.
[27] Kieffer M, Master V, Waites R, et al. TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis[J]. The Plant Journal for Cell & Molecular Biology, 2011, 68(1): 147−158.
[28] 张春雷. 拟南芥TCP15、TCP22基因功能的研究[D]. 兰州: 兰州大学, 2012. Zhang C L. Study on the function of TCP15 and TCP22 genes in Arabidopsis thaliana[D]. Lanzhou: Lanzhou University, 2012.
[29] 冯志娟, 徐盛春, 刘娜, 等. 植物TCP转录因子的作用机理及其应用研究进展[J]. 植物遗传资源学报, 2018, 19(1):112−121. Feng Z J, Xu S C, Liu N, et al. Molecular mechanisms and applications of TCP transcription factors in plants[J]. Journal of Plant Genetic Resources, 2018, 19(1): 112−121.
[30] Mukhopadhyay P, Tyagi A K, Tyagi A K. OsTCP19 influences developmental and abiotic stress signaling by modulating ABI4-mediated pathways[J/OL]. Scientific Reports, 2015, 5: 9998 [2018−11−30]. https://doi.org/10.1038/srep09998.
[31] Li S, Zachgo S. TCP3 interacts with R2R3-MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana[J]. The Plant Journal, 2013, 76(6): 901−913. doi: 10.1111/tpj.2013.76.issue-6
[32] Xiaoyan W, Jiong G, Zheng Z, et al. TCP transcription factors are critical for the coordinated regulation of ISOCHORISMATE SYNTHASE 1 expression in Arabidopsis thaliana[J]. The Plant Journal, 2015, 82(1): 151−162. doi: 10.1111/tpj.2015.82.issue-1
[33] 刘春浩, 梁楠松, 于磊, 等. 水曲柳TCP4转录因子克隆及胁迫和激素下的表达分析[J]. 北京林业大学学报, 2017, 39(6):22−31. Liu C H, Liang N S, Yu L, et al. Cloning of TCP4 transcription factor from ash and expression analysis under stress and hormone[J]. Journal of Beijing Forestry University, 2017, 39(6): 22−31.
[34] Lei N, Yu X, Li S, et al. Phylogeny and expression pattern analysis of TCP transcription factors in cassava seedlings exposed to cold and/or drought stress[J]. Scientific Reports, 2017, 7(1): 10016. doi: 10.1038/s41598-017-09398-5
[35] 杨洋. 白桦BpTCP7基因的功能研究[D]. 哈尔滨: 东北林业大学, 2016. Yang Y. Function analysis of BpTCP7 gene in Betula platyphylla[D]. Harbin: Northeast Forestry University, 2016.
[36] Viola I L, Camoirano A, Gonzalez D H. Redox-dependent modulation of anthocyanin biosynthesis by the TCP transcription factor TCP15 during exposure to high light intensity conditions in Arabidopsis[J]. Plant Physiology, 2015, 34(1): 35−40.
-
期刊类型引用(2)
1. 魏红洋,张一帆,董灵波,刘兆刚,陈莹. 帽儿山主要林分类型空间结构状态综合评价. 中南林业科技大学学报. 2021(10): 131-139 .  百度学术
百度学术
2. 刘文桢,袁一超,张连金,赵中华. 基于林分内部状态与邻域环境的油松林稳定性评价. 林业科学. 2021(09): 76-86 .  百度学术
百度学术
其他类型引用(3)



 下载:
下载: