EST-SSR analysis of genetic diversity of Robinia pseudoacacia clones in Jixian County, Shanxi Province of northern China
-
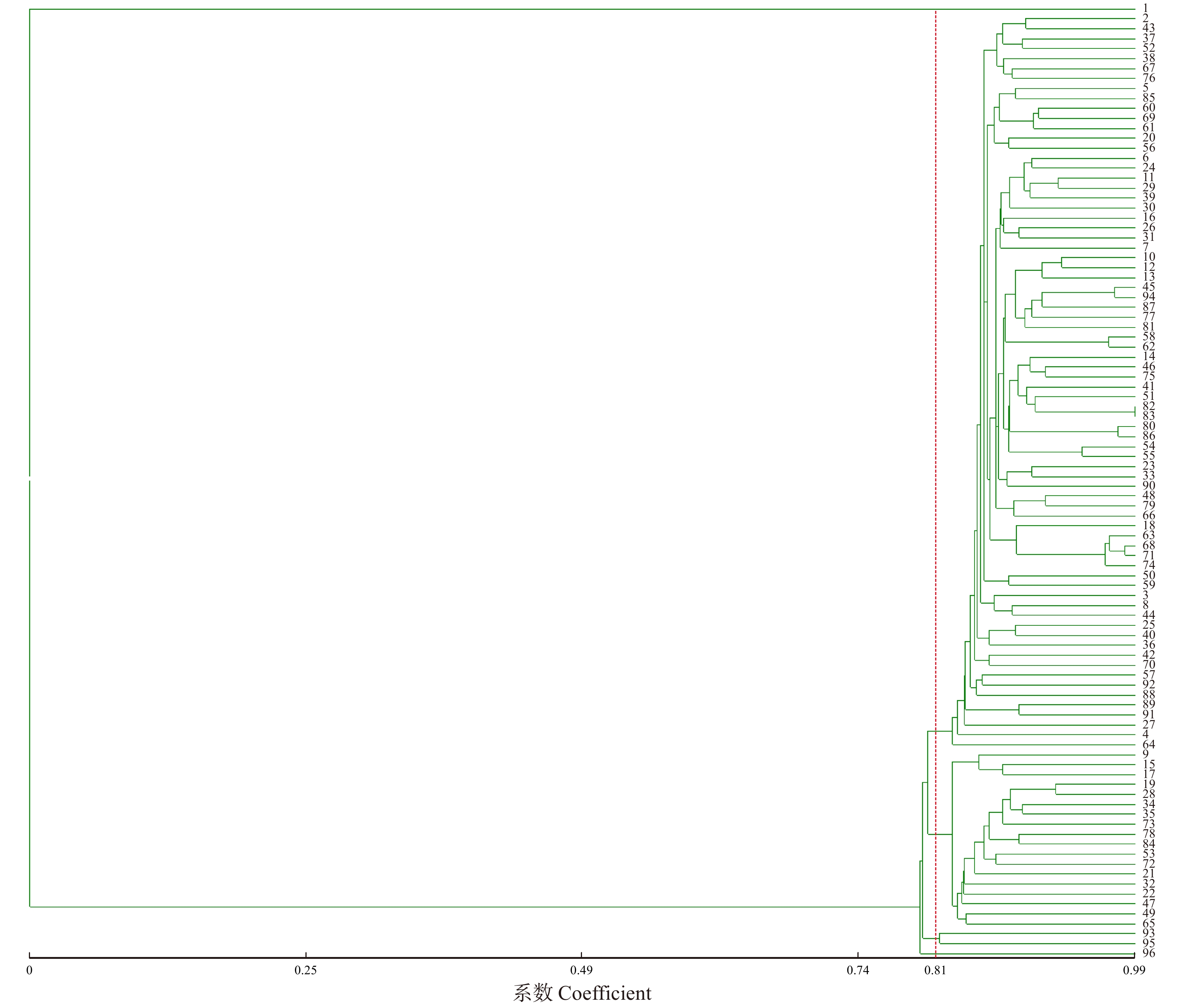
摘要:目的了解各对EST-SSR引物检出的等位简单序列重复片段数量,解析吉县良种基地刺槐无性系种质的多态性状况与种质间亲缘关系,为引物利用和刺槐种质遗传管理提供参考。方法利用课题组开发的45对EST-SSR引物进行PCR扩增,使用毛细管电泳仪对产物进行检测,用Popgene 1.32版软件对群体遗传参数进行评估,利用NTsys 2.10e进行聚类分析。结果45对EST-SSR引物中,除了引物RP-24均表现出良好的多态性。44对引物共检测到等位重复序列298个,每对引物检测到2 ~ 15个,平均6.77个。群体观测杂合度(Ho)的变化范围为0.022 7 ~ 0.842 1,平均为0.412 6。期望杂合度(He)的变化范围为0.088 7 ~ 0.822 3,平均为0.484 0。引物多态性信息量(PIC)变化范围为0.085 2 ~ 0.796 6,平均为0.444 4。结论该基地的刺槐无性系种质具有较高的遗传多样性。由聚类结果可知,在遗传距离为0.81时,可将96份刺槐无性系种质聚成5类。聚类图在分子水平上显示了刺槐无性系种质间的亲缘关系,研究结果对于利用这些引物用于刺槐的种质评价、资源管理和辅助育种有积极的参考价值。Abstract:Objective This paper aims to understand the number of alleles of simple sequence repeats detected by EST-SSR primers, and to analyze the polymorphisms of germplasm of Robinia pseudoacacia clones in Ji xian County of Shanxi Province,northern China and the genetic relationship between germplasms, and provide reference for primer utilization and genetic management of Robinia pseudoacacia germplasm.Method 45 pairs of EST-SSR primers developed by our laboratory were used for PCR amplification, and the products were detected by capillary electrophoresis, the genetic parameters of population were evaluated by Popgene 1.32 software, and cluster analysis was performed by NTsys 2.10e.Result Except for primer RP-24, 45 pairs of EST-SSR primers showed good polymorphism. A total of 298 allelic repeat sequences were detected in 44 pairs of primers, and 2 to 15 were detected in each pair, with an average of 6.77. The observed heterozygosity (Ho) of the population ranged from 0.022 7 to 0.842 1, with an average of 0.412 6. The expected heterozygosity (He) varies from 0.088 7 to 0.822 3, averaging 0.484 0. Primer polymorphism information (PIC) ranged from 0.085 2 to 0.796 6, with an average of 0.444 4.Conclusion Robinia pseudoacacia clonal germplasm in this base has high genetic diversity. It can be seen from the clustering results that 96 genetic clones of the hedgehog clones can be clustered into five classes when the genetic distance is 0.81. Cluster maps showed the genetic relationship among the clones of Robinia pseudoacacia at the molecular level, and the results of the study have positive reference value for the use of these primers for germplasm evaluation, resource management and assisted breeding of Robinia pseudoacacia.
-
Keywords:
- Robinia pseudoacacia /
- EST-SSR /
- genetic diversity /
- cluster analysis
-
-
表 1 试验所用EST-SSR引物信息
Table 1 Information of EST-SSR primers used in the experiment
位点 Locus 引物序列 Primer sequence (5′−3′) 重复基元 Repeat motif 大小 Size/bp 退火温度 Ta/℃ Rp-01 F: TGCAGAAAGAGAAAGCAGAGG (TGTGAA)4 140 58 R: CCGAACCCTTTCTGGTTAGTC Rp-02 F: GCTGCGTTTAATTTTGTCAGG (GAAT)4 170 58 R: TCAATCCATCAAAGAGGAAACA Rp-03 F: GTGAGAAGTGGTTAGGGTTTT (CTC)7 186 55 R: TCAAGATCACCAACGTACAA Rp-04 F: CTCGTGATGATGGTGTTGATG (AATGGT)4 146 58 R: AATGGTCCAAACAACACGAAG Rp-05 F: CCTTGCACATTTATCCCAGAA (TCTGGC)3 158 57 R: CGACCTCGATCTTTTCTTGTG Rp-06 F: TGGACAAAACATCATCGTGTG (TGAGTT)4 147 59 R: CTCTCTTCTTTCTGCCCCTCA Rp-07 F: TTTTTCTCCCAACGAAACAAA (CT)10 144 56 R: TGATGTGTTGTACGGAGGTGA Rp-08 F: TCAGGTGCATAAGCTCATTACTTC (AAAAT)4 152 56 R: GGTTGTCAGATGAAATGCACA Rp-09 F: CGTTTAGAAGCTGAGGCAGAA (CTTT)5 153 58 R: TGAGATATCTTAGTGCAGGAGCA Rp-10 F: GGCATGTGGCTATGAAGATGT (CCTTT)4 154 57 R: TCAGTGGGACTTGGTTTCTTG Rp-11 F: GAAGCTATCACCGCAAATGAA (AG)10 150 57 R: GTCGAAGTGCGTCCTAGATCA Rp-12 F: AAGAGTCATCACGGAGACCAA (AGCAGA)4 150 56 R: GGAGTCCAATTAAGTGCGAGA Rp-13 F: CATTTCCGATTTCCAATTCCT (CTCTTC)4 151 56 R: GCCGAGGACTCGGTAGAAGT Rp-14 F: TTAGCACGAACCTGGTTATGG (TGCAAC)4 151 56 R: CACTTCATTTGGTTCCTTGAGA Rp-15 F: TTAACTAATGCGGCGAGAAGA (TCAC)5 119 56 R: GAGAGGAAGTGTGCGAAACAA Rp-16 F: TATGAGACAGTGTTGGTTGGT (TTCAGT)4 175 56 R: CGTGCCAGAAGAGTATAACAG Rp-17 F: GTAAGTCTGCAAAGAAGACCA (AACCA)4 150 56 R: GCTTTTCACCTATCAACTCAA Rp-18 F: GGATGAACTTTGGCAATCCTT (GGTCAG)4 158 55 R: AATTTGTTGGGAATGCTGTTG Rp-19 F: CAGGAGTGGCAGCATTAGTGT (AGGCTG)4 123 56 R: CACAACAAGCACATTTTGCAC Rp-20 F: TTTCTTGGCTTGCTTTTGCTA (GCAGCT)3 145 56 R: TCTTGGATACGCAAGGTTGTC Rp-21 F: TATGATCACGTCCCCTAATGC (CCA)7 146 57 R: AAGTGGAAAGAAATGGGATGG Rp-22 F: GGTAAGGTGAAGGAGGTGGAG (AGGGTT)4 150 56 R: AGCTTGGTCTCCTAGGTCGTC Rp-23 F: GGAGGAGCAACCATCTGTGTA (AGAAGT)4 146 56 R: CTCCCTCTTCATCCTCACCTC Rp-24 F: TGCACATATTTGCCTGGTTTA (AATA)4 160 56 R: AAAATGAGCATGACACAACCA Rp-25 F: CGGCAACAAGTTGAGAAGAAC (AAAG)5 139 56 R: GGCTCACAAACCAACCTATGA Rp-26 F: GCTGCAAGCAAAGGATCTTAC (ATGATA)4 139 56 R: CCTCATCATCCTCGTCATCAT Rp-27 F: TGGACAAAACATCATCGTGTG (TGAGTT)4 147 57 R: CTCTCTTCTTTCTGCCCCTCA Rp-28 F: CTTGGTCTAGAAAGTCCTGCT (CAG)7 151 56 R: GGTCATCAAGGTTAGTTGGAT Rp-29 F: CCTGATGATCAAAACGACGAC (GATC)4 148 56 R: GGAGGTGACCCCTCTTATCCT Rp-30 F: TTGAACCAAAACTGGAAGAGC (GCT)8 151 56 R: GCACCGTACAGTTACCCTATCC Rp-31 F: GACCCCATTTTTCTCAAGGAC (ATT)7 140 56 R: TTGGATAAGTCGGTGAAGGTG Rp-32 F: CCACGTGGTTCTTCAAACATT (GTG)7 163 57 R: CAACAACAACCCACAAACACA Rp-33 F: CAAACAGTCTCATGGAAATGGA (ATC)7 141 56 R: GGGTTGGTATTGTTGGGAAAT Rp-34 F: AGGATATTAGCCAAGTCCATC (TGGTGA)4 164 57 R: AGTAACCATCACCACAATCAC Rp-35 F: TCAGACGTGGTAGAGCAGTGTT (CACAC)4 152 58 R: ATTTGTTTTTGGGGGAGATTG Rp-36 F: CGTTTCAGCCATTGATTTTGT (GAATC)5 141 57 R: GATCATCACCGTCCACCTTC Rp-37 F: TGTCGTCATTTTATTTTACCC (GAACGA)4 152 56 R: CTCACCCTTTTTATTTCCATT Rp-38 F: TCCATTCCCTGGTTTCTTCTT (TC)10 150 56 R: AGCACAATTTCCTCAGTGCAG Rp-39 F: TTAAAGAATGTTCCGTTCAGA (AAGAGG)3 152 56 R: GAGAAGATAGCCTCCTAGCTG Rp-40 F: TCATTGGACATCCCTCCATAA (TAA)8 139 56 R: GGCTCGACATGGTTGATTTT Rp-41 F: AACTCACCCAATTGCACACTC (CCA)7 143 56 R: GAGCAAGAGCTAAAGCAGCAA Rp-42 F: CTTCGCAATCCTCACTCTTTG (AATC)4 169 55 R: CTTACCCAGAAGCCAACAATG Rp-43 F: CAAAGCAGAGAGAATGTATGG (CAAAAT)4 155 57 R: ATCCCTTGCTCCTTGTAATAG Rp-44 F: TATCTGGGAGAATCGAGAGCA (ATCA)5 145 57 R: CCACCATGGTTGTCCTTCTAA Rp-45 F: GGGTTGAGGAAGAGAGGAGAA (TTC)7 156 57 R: AAAAATCGAATCGTGTTGGTG 表 2 44对引物扩增结果及多态性信息
Table 2 Amplification results and polymorphic information of 44 pairs of primers
引物 Primer 观测杂合度 Ho 期望杂合度 He Nei氏指数 Nei 等位重复序列 Na 有效等位重复序列 Ne Shannon信息指数 I 引物多态性信息量 PIC RP-01 0.623 7 0.629 4 0.626 0 6 2.674 0 1.152 8 0.557 1 RP-02 0.106 7 0.125 3 0.124 4 2 1.142 1 0.244 9 0.116 8 RP-03 0.641 3 0.628 1 0.624 6 11 2.664 1 1.329 8 0.571 7 RP-04 0.781 6 0.790 8 0.786 2 8 4.678 0 1.638 4 0.752 4 RP-05 0.540 2 0.673 0 0.669 2 15 3.022 8 1.680 0 0.652 5 RP-06 0.734 0 0.738 4 0.734 4 7 3.765 6 1.445 5 0.690 5 RP-07 0.842 1 0.816 8 0.811 5 11 5.303 9 1.892 0 0.788 2 RP-08 0.505 4 0.726 6 0.722 7 7 3.606 8 1.426 1 0.679 1 RP-09 0.648 9 0.660 8 0.657 3 15 2.918 1 1.529 9 0.615 7 RP-10 0.433 0.422 9 0.420 6 5 1.725 8 0.798 4 0.375 7 RP-11 0.719 1 0.822 3 0.817 7 14 5.485 5 2.018 3 0.796 6 RP-12 0.750 0 0.776 8 0.772 7 10 4.400 1 1.646 2 0.737 7 RP-13 0.315 8 0.362 2 0.360 3 9 1.563 2 0.878 1 0.350 0 RP-14 0.395 6 0.708 7 0.704 8 10 3.387 6 1.556 5 0.671 7 RP-15 0.416 7 0.371 6 0.369 6 8 1.586 4 0.802 4 0.345 2 RP-16 0.162 8 0.173 2 0.172 2 4 1.208 0 0.374 9 0.163 6 RP-17 0.250 0 0.247 7 0.246 4 7 1.327 0 0.572 3 0.236 1 RP-18 0.434 2 0.599 3 0.595 4 7 2.471 5 1.085 8 0.512 3 RP-19 0.446 8 0.610 6 0.607 4 7 2.547 1 1.118 6 0.528 9 RP-20 0.046 0 0.088 7 0.088 2 3 1.096 7 0.203 8 0.085 2 RP-21 0.246 8 0.280 6 0.278 8 6 1.386 6 0.624 5 0.266 4 RP-22 0.602 2 0.595 9 0.592 7 5 2.455 0 1.141 4 0.546 7 RP-23 0.341 2 0.502 1 0.499 1 6 1.996 4 0.957 2 0.446 5 RP-25 0.104 2 0.139 2 0.138 5 5 1.160 7 0.345 2 0.134 9 RP-26 0.212 8 0.214 0 0.212 8 6 1.270 4 0.472 0 0.201 0 RP-27 0.118 3 0.112 5 0.111 9 3 1.126 0 0.242 6 0.106 6 RP-28 0.651 7 0.719 5 0.715 4 7 3.514 2 1.436 0 0.671 6 RP-29 0.022 7 0.107 8 0.107 2 2 1.120 0 0.218 1 0.101 4 RP-30 0.268 8 0.416 6 0.414 3 5 1.707 4 0.775 8 0.367 3 RP-31 0.376 6 0.561 2 0.557 6 10 2.260 4 1.269 3 0.532 9 RP-32 0.511 9 0.635 9 0.632 1 7 2.718 0 1.246 2 0.578 3 RP-33 0.557 9 0.444 1 0.441 7 4 1.791 2 0.696 4 0.357 3 RP-34 0.500 0 0.523 5 0.520 6 8 2.086 0 1.106 7 0.485 3 RP-35 0.375 0 0.565 1 0.561 9 5 2.282 7 0.987 2 0.486 1 RP-36 0.512 2 0.661 5 0.657 4 4 2.919 0 1.167 6 0.590 9 RP-37 0.612 9 0.665 1 0.661 5 6 2.954 4 1.353 5 0.628 0 RP-38 0.556 8 0.774 9 0.770 5 15 4.357 9 1.895 0 0.741 3 RP-39 0.392 4 0.592 3 0.588 5 5 2.430 3 1.021 3 0.507 8 RP-40 0.178 9 0.182 0 0.181 0 3 1.221 0 0.345 7 0.166 4 RP-41 0.315 8 0.331 3 0.329 5 6 1.491 5 0.706 5 0.311 7 RP-42 0.281 7 0.459 5 0.456 3 2 1.839 1 0.648 7 0.352 2 RP-43 0.221 1 0.305 3 0.303 7 4 1.436 1 0.613 7 0.285 1 RP-44 0.081 4 0.133 3 0.132 6 4 1.152 8 0.317 1 0.129 0 RP-45 0.315 2 0.398 4 0.396 2 4 1.656 2 0.661 0 0.333 4 总计 Total 298 104.907 6 均值 Mean 0.412 6 0.484 0 0.481 2 6.772 7 2.384 3 0.991 9 0.444 4 -
[1] Huntley J C. Robinia pseudoacacia L. black locust[M]. Washington DC: Silvics of North America, 1990, 2: 755−761.
[2] 赵蓬晖, 张江涛, 王念. 刺槐原产地分布及世界各国引种与研究概况[J]. 河南林业科技, 2017, 37(1):30−32. doi: 10.3969/j.issn.1003-2630.2017.01.010 Zhao P H, Zhang J T, Wang N. The original distribution introduction and development of Robinia pserdoacacia[J]. Journal of Henan Forestry Science and Technology, 2017, 37(1): 30−32. doi: 10.3969/j.issn.1003-2630.2017.01.010
[3] 中国森林编辑委员会. 中国森林: 3卷[M]. 北京: 中国林业出版社, 2000: 3−5. China Forest Editorial Committee. Forests in China: Vol. 3[M]. Beijing: China Forestry Publishing House, 2000: 3−5.
[4] 孙鹏, 戴丽, 孙妍, 等. 刺槐幼林不同无性系性状比较及相关分析[J]. 北京林业大学学报, 2013, 35(4):34−40. Sun P, Dai L, Sun Y, et al. Comparison and correlative analysis of growth traits in young Robinia pseudoacacia clones[J]. Journal of Beijing Forestry University, 2013, 35(4): 34−40.
[5] 习洋, 胡瑞阳, 王欢, 等. 刺槐未成熟合子胚的体细胞胚胎发生和植株再生[J]. 林业科学, 2012, 48(1):60−69. doi: 10.3969/j.issn.1672-8246.2012.01.009 Xi Y, Hu R Y, Wang H, et al. Somatic embryogenesis and plant regeneration from immature zygotic embryos of black locust (Robinia pseudoacacia)[J]. Scientia Silvae Sinicae, 2012, 48(1): 60−69. doi: 10.3969/j.issn.1672-8246.2012.01.009
[6] 沈俊岭, 赵芳, 李云, 等. 速生型刺槐遗传转化体系的建立[J]. 核农学报, 2006, 20(6):477−481. doi: 10.3969/j.issn.1000-8551.2006.06.007 Shen J L, Zhao F, Li Y, et al. Establishment of genetic transformation system of fastgrowing black locust[J]. Acta Agriculturae Nucleatae Sinica, 2006, 20(6): 477−481. doi: 10.3969/j.issn.1000-8551.2006.06.007
[7] 孙鹏, 戴丽, 徐兆翮, 等. 刺槐花粉萌发和花粉管伸长生长过程中柱头与花柱内钙离子的分布特征[J]. 北京林业大学学报, 2015, 37(11):59−68. Sun P, Dai L, Xu Z H, et al. Distribution of Ca2+ in the stigma and style of Robinia pseudoacacia during pollen germination and pollen tube elongation[J]. Journal of Beijing Forestry University, 2015, 37(11): 59−68.
[8] Powell W, Machray G C, Provan J. Polymorphism revealed by simple sequence repeats[J]. Trends in Plant Science, 1996, 1(7): 215−222.
[9] 唐荣华, 张君诚, 吴为人. SSR分子标记的开发技术研究进展[J]. 西南农业学报, 2002, 15(4):106−109. doi: 10.3969/j.issn.1001-4829.2002.04.026 Tang R H, Zhang J C, Wei W R. Progress in the way to develop SSR molecular marker[J]. Southwest China Journal of Agricultural Sciences, 2002, 15(4): 106−109. doi: 10.3969/j.issn.1001-4829.2002.04.026
[10] 李卫国, 常天俊, 龚红梅. EST-SSR及其在植物基因组学研究中的应用[J]. 生物技术, 2008, 18(4):90−93. Li W G, Chang T J, Gong H M. Simple sequence repeats derived from expression sequence tags (EST-SSRs) and its application to plant genomics[J]. Biotechnology, 2008, 18(4): 90−93.
[11] 陈全求, 詹先进, 蓝家样, 等. EST分子标记开发研究进展[J]. 中国农学通报, 2008, 24(9):72−77. Chen Q Q, Zhan X J, Lan J Y, et al. Study progress in development of EST(expressed sequence tags)[J]. Chinese Agricultural Science Bulletin, 2008, 24(9): 72−77.
[12] 徐杨, 邓丽丽, 周丽, 等. 云南松EST-SSR引物在其近缘种中通用性的研究[J]. 西南林业大学学报, 2016, 36(1):16−20. Xu Y, Deng L L, Zhou L, et al. The transferability analysis of microsatellite markers from expressed sequence tags of Pinus yunnanensis to its close related species[J]. Journal of Southwest Forestry College, 2016, 36(1): 16−20.
[13] Varshney R K, Graner A, Sorrells M E. Genic microsatellite markers in plants: features and applications[J]. Trends in Biotechnology, 2005, 23(1): 48−55. doi: 10.1016/j.tibtech.2004.11.005
[14] 林元震, 郭海, 刘纯鑫, 等. EST-SSR标记在木本植物中的开发和应用[J]. 植物生理学通讯, 2009, 45(12):1221−1225. Lin Y Z, Guo H, Liu C X, et al. Development and application of EST-SSR markers in woody plants[J]. Plant Physiology Communications, 2009, 45(12): 1221−1225.
[15] 赵天梁. 山西华北落叶松群落物种多样性[J]. 北京林业大学学报, 2017, 39(6):45−50. Zhao T L. Species diversity of Larix principis-rupprechtii communities in Shanxi Province of northern China[J]. Journal of Beijing Forestry University, 2017, 39(6): 45−50.
[16] 安宗燕, 唐红, 李婉茹. 基于EST-SSR的紫斑牡丹品种遗传多样性分析[J]. 分子植物育种, 2018, 16(20):6744−6752. An Z Y, Tang H, Li W R. Genetic diversity analysis of Paeonia rockii cultivar based on EST-SSR[J]. Molecular Plant Breeding, 2018, 16(20): 6744−6752.
[17] 石艳, 童再康, 高燕会. 换锦花EST-SSR标记开发及遗传多样性分析[J]. 核农学报, 2018, 32(6):1089−1096. doi: 10.11869/j.issn.100-8551.2018.06.1089 Shi Y, Tong Z K, Gao Y H. Development of EST-SSR markers and genetic diversity analysis in Lycoris sprengeri[J]. Acta Agriculturae Nucleatae Sinica, 2018, 32(6): 1089−1096. doi: 10.11869/j.issn.100-8551.2018.06.1089
[18] 胡文舜, 陈秀萍, 郑少泉. 龙眼EST-SSR标记开发及无患子科5个属种质遗传多样性分析[J/OL]. 园艺学报, 2019 (2019−01−05) [2019−02−13]. http://kns.cnki.net/kcms/detail/11.1924.S.20190103.1338.006.html. Hu W S, Chen X P, Zheng S Q. EST-SSR markers development from Longan (Dimocarpus longan Lour.) and its application in genetic diversity analysis of five genera of Sapindaceae[J/OL]. Acta Horticulturae Sinica, 2019 (2019−01−05) [2019−02−13]. http://kns.cnki.net/kcms/detail/11.1924.S.20190103.1338.006.html.
[19] 王西成, 姜淑苓, 上官凌飞, 等. 梨EST-SSR标记的开发及其在梨品种遗传多样性分析中的应用评价[J]. 中国农业科学, 2010, 43(24):5079−5087. doi: 10.3864/j.issn.0578-1752.2010.24.012 Wang X C, Jiang S L, Shangguan L F, et al. Development of EST-SSR markers for pear and evaluation of their application in pear genetic diversity analysis[J]. Scientia Agricultura Sinica, 2010, 43(24): 5079−5087. doi: 10.3864/j.issn.0578-1752.2010.24.012
[20] 胡宏霞, 穆国俊, 侯名语, 等. 河北省花生地方品种基于EST-SSR的遗传多样性及性状−标记相关分析[J]. 植物遗传资源学报, 2013, 14(6):1118−1123, 1129. Hu H X, Mu G J, Hou M Y, et al. Genetic diversity and marker-trait analysis for the peanut Landraces in Hebei based on EST-SSR markers[J]. Journal of Plant Genetic Resources, 2013, 14(6): 1118−1123, 1129.
[21] 王盈盈, 刘玉新, 汪俊君, 等. 62个小麦品种基于EST-SSR标记的遗传多样性分析[J]. 麦类作物学报, 2008, 28(5):749−754. Wang Y Y, Liu Y X, Wang J J, et al. Genetic diversity of 62 commom wheat cultivars using EST-SSR markers[J]. Journal of Triticeae Crops, 2008, 28(5): 749−754.
[22] 赵克奇, 董黎, 王少明, 等. 刺槐EST-SSR标记PCR反应体系的优化[J]. 中国农学通报, 2014, 30(22):45−52. doi: 10.11924/j.issn.1000-6850.2014-0325 Zhao K Q, Dong L, Wang S M, et al. The optimization of EST-SSR PCR reaction system for Robinia pseudoacacia L.[J]. Chinese Agricultural Science Bulletin, 2014, 30(22): 45−52. doi: 10.11924/j.issn.1000-6850.2014-0325
[23] Guo Q, Wang J X, Su L Z, et al. Development and Evaluation of a Novel Set of EST-SSR Markers Based on Transcriptome Sequences of Black Locust (Robinia pseudoacacia L.)[J/OL]. Genes, 2017, 8(7) (2017−07−07) [2017−12−15]. https://doi.org/10.3390/genes8070177.
[24] 尹明华, 徐文慧, 谢妮妮, 等. 三叶青种质资源遗传多样性的SSR荧光标记分析[J]. 中草药, 2018, 49(23):5649−5656. doi: 10.7501/j.issn.0253-2670.2018.23.026 Yin M H, Xu W H, Xie N N, et al. Genetic diversity analysis of Tetrastigma hemsleyanum germplasm resources based on fluorescently labeled SSR markers[J]. Chinese Traditional and Herbal Drugs, 2018, 49(23): 5649−5656. doi: 10.7501/j.issn.0253-2670.2018.23.026
[25] Selkoe K A, Toonen R J. Microsatellites for ecologists: a practical guide to using and evaluating microsatellite markers[J]. Ecology Letters, 2006, 9(5): 615−629. doi: 10.1111/ele.2006.9.issue-5
[26] Guo Q, Li, X Y, Yang S H, et al. Evaluation of the genetic diversity and differentiation of black locust (Robinia pseudoacacia L.) based on genomic and expressed sequence Tag-simple sequence repeats[J]. Moleculai Sciences, 2018, 19(9): 2492.
[27] Botstein D. Construction of a genetic linkage map in man using restriction fragment length polymorphisms[J]. Am J Hum Genet, 1980, 32(3): 314−331.
[28] Pandian S, Satish L, Rameshkumar R, et al. Analysis of population structure and genetic diversity in an exotic germplasm collection of Eleusine coracana (L.) Gaertn. using genic-SSR markers[J]. Gene, 2018, 653: 80−90.
[29] 毛秀红, 郑勇奇, 孙百友, 等. 基于SSR的刺槐无性系遗传多样性分析和指纹图谱构建[J]. 林业科学, 2017, 53(10):80−89. doi: 10.11707/j.1001-7488.20171009 Mao X H, Zheng Y Q, Sun B Y, et al. Genetic diversity and fingerprints of Robinia pseudoacacia clones based on SSR markers[J]. Scientia Silvae Sinicae, 2017, 53(10): 80−89. doi: 10.11707/j.1001-7488.20171009
[30] 孙芳, 杨敏生, 张军, 等. 刺槐不同居群遗传多样性的ISSR分析[J]. 植物遗传资源学报, 2009, 10(1):91−96. Sun F, Yang M S, Zhang J, et al. ISSR analysis of genetic diversity of Robinia pseudoacacia populations[J]. Journal of Plant Genetic Resources, 2009, 10(1): 91−96.
[31] 王东升, 周继磊, 解荷锋, 等. 刺槐无性系遗传多样性的AFLP分析[J]. 西南林业大学学报, 2012, 32(6):25−29. doi: 10.3969/j.issn.2095-1914.2012.06.005 Wang D S, Zhou J L, Xie H F, et al. AFLP genetic diversity analysis of Robinia pseudoacacia clones[J]. Journal of Southwest Forestry College, 2012, 32(6): 25−29. doi: 10.3969/j.issn.2095-1914.2012.06.005
[32] 杨敏生, Hertel H,Schneck V. 欧洲刺槐种源群体遗传结构和多样性[J]. 生态学报, 2004, 24(12):2700−2706. doi: 10.3321/j.issn:1000-0933.2004.12.004 Yang M S,Hertel H,Schneck V. Genetic diversity and population structure of Robinia pseudoacacia provenances from middle Europe[J]. Acta Ecologica Sinica, 2004, 24(12): 2700−2706. doi: 10.3321/j.issn:1000-0933.2004.12.004
[33] 张志毅, 林木遗传学基础[M]. 北京: 中国林业出版社, 2012: 304 Zhang Z Y, Genetic basis of forest trees[M]. Beijing: China Forestry Publishing House, 2012: 304
[34] 骆蒙, 贾继增. 植物基因组表达序列标签(EST)计划研究进展[J]. 生物化学与生物物理进展, 2001, 28(4):494−497. doi: 10.3321/j.issn:1000-3282.2001.04.014 Luo M, Jia J Z. Progress in expressed sequence tags (EST) project of plant genome[J]. Progress in Biochemistry and Biophysics, 2001, 28(4): 494−497. doi: 10.3321/j.issn:1000-3282.2001.04.014
-
期刊类型引用(0)
其他类型引用(4)



 下载:
下载:
