Cloning the promoter of BpSPL8 from Betula platyphylla and overexpression of BpSPL8 gene affecting drought tolerance in Arabidopsis thaliana
-
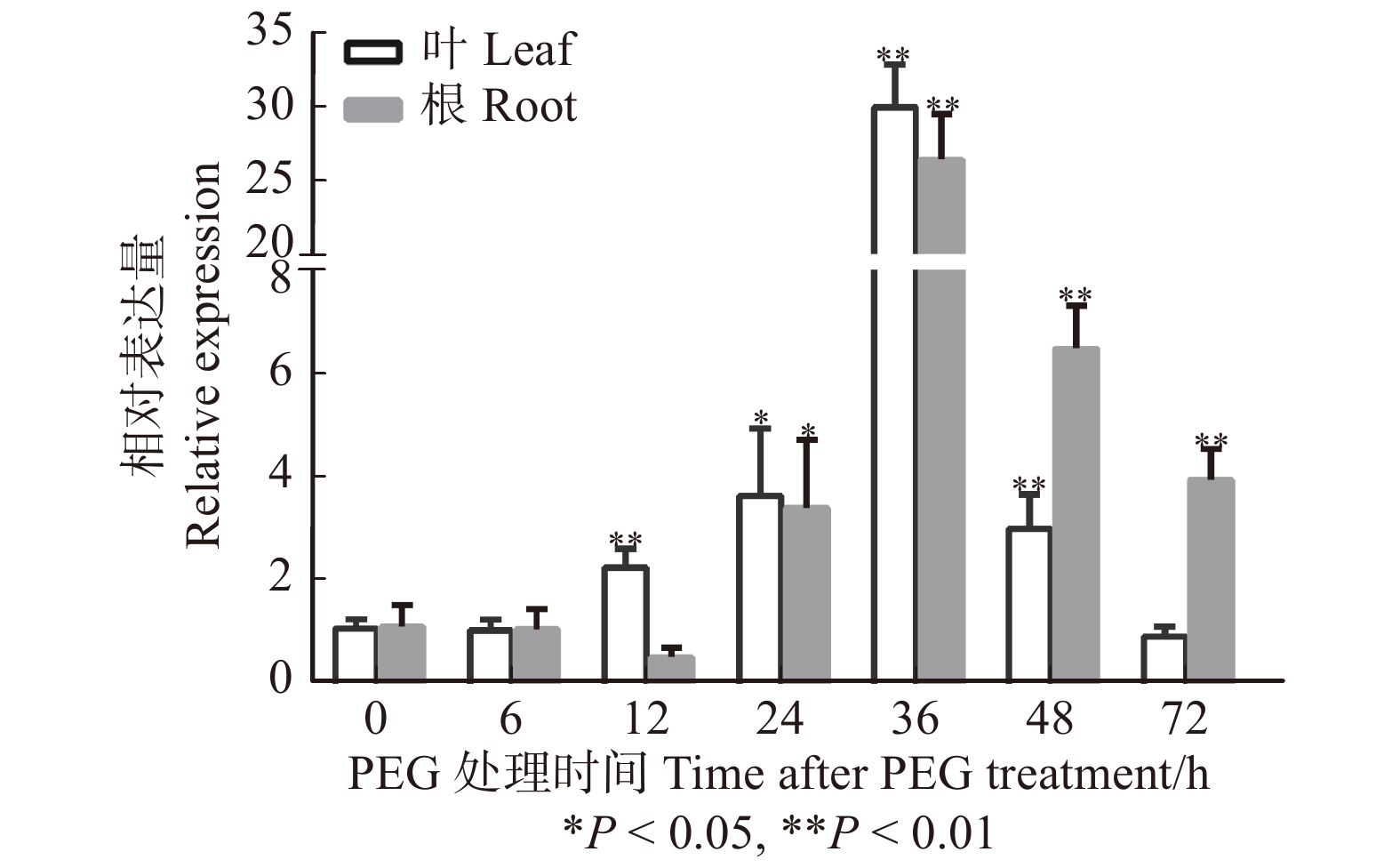
摘要:目的目前对植物SPL8的基因功能研究主要集中在开花和育性方面,而其在干旱胁迫响应中的作用却鲜有报道。本文克隆、分析了白桦BpSPL8启动子,并研究了BpSPL8基因在拟南芥中响应干旱胁迫的功能。方法通过PCR克隆技术得到了白桦BpSPL8启动子;利用PLACE和PlantCARE软件对BpSPL8启动子顺式作用元件进行了预测。构建了BpSPL8启动子驱动GUS(β-葡萄糖苷酸酶编码基因)的植物表达载体,并采用浸花法将其转化至拟南芥中;继而利用GUS染色分析了BpSPL8启动子的组织表达模式;同时对BpSPL8在PEG处理下的表达水平进行了qRT-PCR分析。最后,以过表达BpSPL8拟南芥为材料来探究BpSPL8在干旱胁迫下的生物学功能。结果启动子元件分析显示,BpSPL8启动子中含有组织特异表达、光响应、激素响应及多个胁迫响应元件。GUS染色结果表明,BpSPL8启动子可在拟南芥的下胚轴、叶片、叶柄、根和花序中启动GUS基因表达。BpSPL8基因在PEG处理下的野生型白桦的根和叶片中均呈现先上调后下调的表达趋势。干旱胁迫下,过表达BpSPL8拟南芥的存活率和脯氨酸含量均显著低于野生型,丙二醛含量显著高于野生型;两个已知的抗逆基因DR29B和P5CS1在干旱处理后的野生型和转基因拟南芥中均上调表达;但在转基因拟南芥中呈现出延迟上调的表达模式。结论异源过表达白桦BpSPL8能够降低拟南芥的耐旱性,并在干旱胁迫下影响抗性基因DR29B和P5CS1的表达模式。Abstract:ObjectiveCurrently, researches on the gene function of plant SPL8 are mainly focused on flowering and fertility, but there are fewer reports about its SPL8 function in drought stress response. In this paper, BpSPL8 promoter was cloned and analyzed from Betula platyphylla, and the function of BpSPL8 gene in response to drought stress was studied in Arabidopsis thaliana.MethodPromoter sequence of BpSPL8 gene was isolated from Betula platyphylla by PCR technology, and the cis-element prediction of BpSPL8 promoter was performed using PLACE and PlantCARE software. The plant expression vectors with GUS (β-glucuronidase coding gene) expression driven by the promoters of BpSPL8 were constructed and transformed into Arabidopsis thaliana by the floral dip method. Through the detection of GUS activity, the tissue expression pattern of the BpSPL8 promoter in Arabidopsis thaliana was analyzed. QRT-PCR analysis was performed on the expression level of BpSPL8 under PEG treatment. Finally, the overexpression of BpSPL8 Arabidopsis thaliana was used to explore the function of BpSPL8 in the drought process.ResultPromoter element analysis revealed that BpSPL8 promoter contained elements for tissue-specific expression, light-responsive, hormone-responsive and stress-responsive. GUS histochemical staining results showed that GUS activity was observed in hypocotyls, leaves, petioles, roots and inflorescences of transgenic Arabidopsis thaliana carrying the BpSPL8 promoter. The expression patterns of BpSPL8 gene in roots and leaves of birch were up-regulated and then down-regulated under PEG treatment. Drought stress tolerance pointed out that the transgenic plants showed significantly lower survival rate and proline content than wild type, while malondialdehyde content was higher than wild type. Two known stress-resistant genes, DR29B and P5CS1, were up-regulated in wild-type and transgenic Arabidopsis thaliana under drought stress. However, compared with wild-type, they showed delayed up-regulation in transgenic Arabidopsis thaliana.ConclusionEctopic overexpression of BpSPL8 can reduce the drought tolerance of Arabidopsis thaliana and affect the expression patterns of resistance genes DR29B and P5CS1 under drought stress.
-
Keywords:
- promoter /
- BpSPL8 /
- Betula platyphylla /
- transgenic /
- drought
-
-
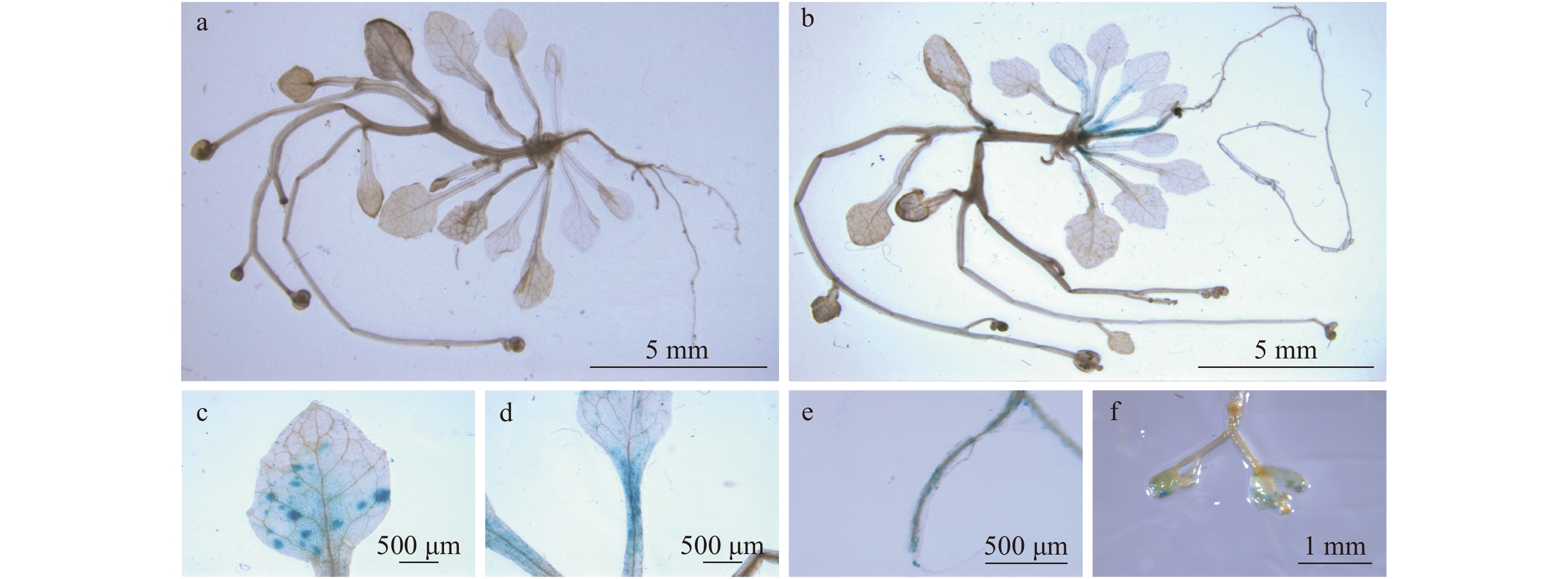
图 1 转ProSPL8::GUS拟南芥的组织化学GUS测定
a. 野生型拟南芥未检测到GUS活性;b. GUS活性在转ProSPL8::GUS拟南芥的下胚轴、叶片、叶柄、根和花序检测到;c. 转ProSPL8::GUS拟南芥叶片;d. 转ProSPL8::GUS拟南芥叶柄;e. 转ProSPL8::GUS拟南芥根;f. 转ProSPL8::GUS拟南芥花序。a, wild type Arabidopsis thaliana without GUS activity; b, GUS activity was observed in hypocotyls, leaves, petioles, roots and inflorescences of ProSPL8::GUS transgenic Arabidopsis thaliana; c, leaves of ProSPL8::GUS; d, petiole of ProSPL8::GUS; e, root of ProSPL8::GUS; f, inflorescence of ProSPL8::GUS.
Figure 1. Histochemical GUS staining of ProSPL8::GUS transgenic Arabidopsis thaliana
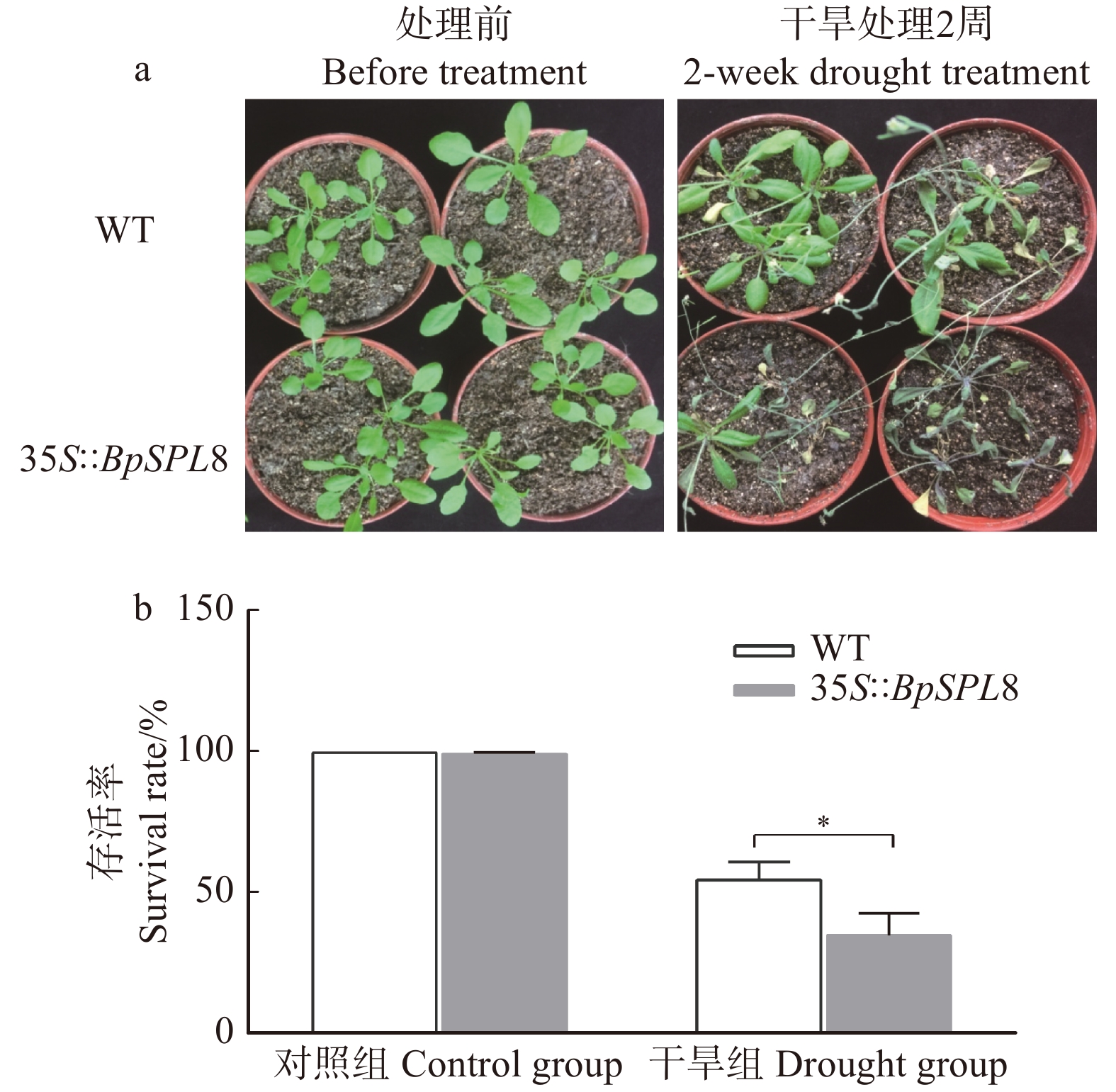
图 3 干旱处理条件下野生型和35S::BpSPL8转基因拟南芥的生长状态及存活率
WT为野生型拟南芥,35S::BpSPL8为BpSPL8过表达拟南芥。a. 处理前和干旱2周后野生型和35S::BpSPL8转基因拟南芥的表型;b. 干旱胁迫2周复水3 d后和正常生长条件下野生型和35S::BpSPL8转基因拟南芥的存活率(*P < 0.05)。WT is wild-type Arabidopsis thaliana, 35S::BpSPL8 is BpSPL8 overexpressing Arabidopsis thaliana. a, phenotype of wild-type and 35S::BpSPL8 transgenic Arabidopsis thaliana before treatment and 2 weeks after drought; b, survival rate of wild-type and 35S::BpSPL8 transgenic Arabidopsis thaliana under normal growth conditions and re-watering 3 days after 2 weeks of drought treatment (*P < 0.05).
Figure 3. Growth status and survival rate of wild-type and 35S::BpSPL8 transgenic Arabidopsis thaliana under drought stress
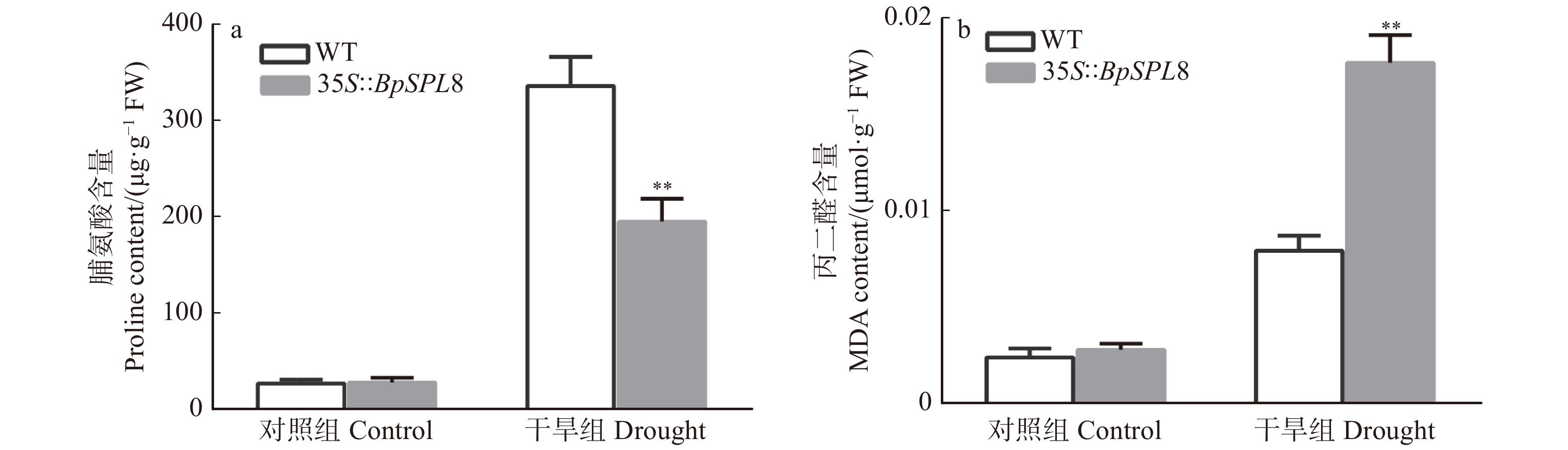
图 4 正常生长和干旱7 d 35S::BpSPL8转基因和野生型拟南芥脯氨酸和丙二醛的含量测定
WT为野生型拟南芥(**P < 0.01),35S::BpSPL8为BpSPL8过表达拟南芥。WT is wild-type Arabidopsis thaliana, 35S::BpSPL8 is BpSPL8 overexpressing Arabidopsis thaliana (**P < 0.01).
Figure 4. Contents of proline and malondialdehyde in wild type and 35S::BpSPL8 transgenic Arabidopsis thaliana under normal growth conditions and 7 days of drought treatment
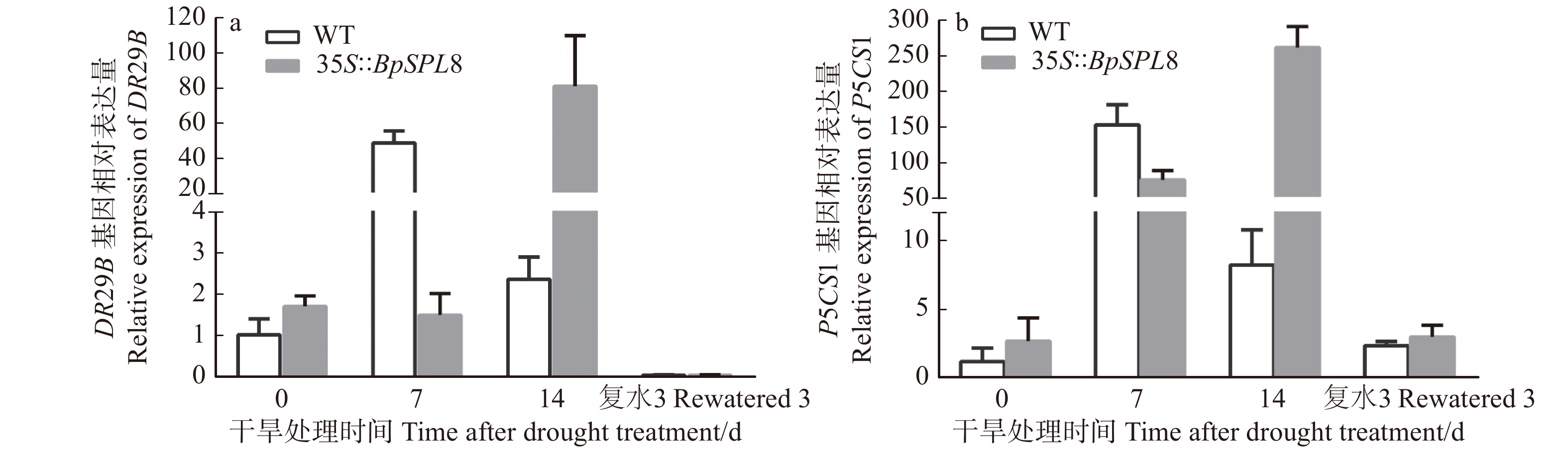
图 5 干旱处理下野生型和35S::BpSPL8转基因拟南芥DR29B和P5CS1表达模式分析
WT为野生型拟南芥;35S::BpSPL8为BpSPL8过表达拟南芥。WT is wild-type Arabidopsis thaliana, 35S::BpSPL8 is BpSPL8 overexpressing Arabidopsis thaliana.
Figure 5. Expression pattern analysis of DR29B and P5CS1 in wild type and
35S::BpSPL8 transgenic Arabidopsis thaliana under drought treatment 表 1 BpSPL8启动子中的顺式作用元件及相关功能预测
Table 1 Cis-acting elements and predicted functions in the sequence of BpSPL8 promoter
顺式元件 Cis-element 数量 Number 功能 Function 功能分类 Function group MBS 4 MYB binding site involved in drought-inducibility Binding site specific response element MRE 1 MYB binding site involved in light responsiveness Binding site specific response element circadian 3 Circadian control Circadian Box-W1 1 Fungal elicitor responsive element Elicitor specific responsive element ABRE 1 Abscisic acid responsiveness Hormone responsive element TGA-element 1 Auxin-responsive element Hormone responsive element GARE-motif 3 Gibberellin-responsive element Hormone responsive element ERE 1 Ethylene-responsive element Hormone responsive element TCA-element 1 Salicylic acid responsiveness Hormone responsive element AE-box 2 Light response Light responsive element Box 4 3 Light response Light responsive element Box I 3 Light responsive element Light responsive element CATT-motif 4 Light responsive element Light responsive element GAG-motif 1 Light responsive element Light responsive element GA-motif 2 Light responsive element Light responsive element GT1-motif 1 Light responsive element Light responsive element H-box 1 Light responsive element Light responsive element Sp1 6 Light responsive element Light responsive element TCT-motif 1 Light responsive element Light responsive element G-Box 2 Light responsiveness Light responsive element G-box 3 Light responsiveness Light responsive element ATGCAAAT motif 1 Associated to the TGAGTCA motif Plant tissue-specific element MSA-like 1 Cell cycle regulation Plant tissue-specific element GCN4_motif 1 Endosperm expression Plant tissue-specific element Skn-1_motif 3 Endosperm expression Plant tissue-specific element CAT-box 1 Meristem expression Plant tissue-specific element AC-II 1 Negative regulation of phloem expression Plant tissue-specific element as-2-box 1 Shoot-specific expression and light responsiveness Plant tissue-specific element ARE 1 Anaerobic response element Stress responsive element TC-rich repeats 1 Defense and stress responsiveness Stress responsive element HSE 2 Heat stress responsiveness Stress responsive element GCC box 1 Wounding and pathogen responsiveness Stress responsive element W box 1 Wounding and pathogen responsiveness Stress responsive element 5UTR Py-rich stretch 2 Conferring high transcription levels Transcription regulation element AAGAA-motif 1 Unknown Unnamed__1 2 Unknown Unnamed__3 2 Unknown Unnamed__4 7 Unknown -
[1] Cardon G, Hohmann S, Klein J, et al. Molecular characterisation of the Arabidopsis SBP-box genes[J]. Gene, 1999, 237(1): 91−104. doi: 10.1016/S0378-1119(99)00308-X
[2] Schwab R, Palatnik J F, Riester M, et al. Specific effects of microRNAs on the plant transcriptome[J]. Developmental Cell, 2005, 8(4): 517−527. doi: 10.1016/j.devcel.2005.01.018
[3] Wu G, Poethig R S. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3[J]. Development, 2006, 133(18): 3539−3547. doi: 10.1242/dev.02521
[4] Wang J W, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana[J]. Cell, 2009, 138(4): 738−749. doi: 10.1016/j.cell.2009.06.014
[5] Yu N, Cai W J, Wang S, et al. Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana[J]. Plant Cell, 2010, 22(7): 2322−2335. doi: 10.1105/tpc.109.072579
[6] Jung J H, Seo P J, Kang S K, et al. miR172 signals are incorporated into the miR156 signaling pathway at the SPL3/4/5 genes in Arabidopsis developmental transitions[J]. Plant Molecular Biology, 2011, 76(1−2): 35−45. doi: 10.1007/s11103-011-9759-z
[7] Shikata M, Koyama T, Mitsuda N, et al. Arabidopsis SBP-box genes SPL10, SPL11 and SPL2 control morphological change in association with shoot maturation in the reproductive phase[J]. Plant and Cell Physiology, 2009, 50(12): 2133−2145. doi: 10.1093/pcp/pcp148
[8] Schwarz S, Grande A V, Bujdoso N, et al. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis[J]. Plant Molecular Biology, 2008, 67(1−2): 183−195. doi: 10.1007/s11103-008-9310-z
[9] Yamasaki H, Hayashi M, Fukazawa M, et al. SQUAMOSA promoter binding protein-like7 is a central regulator for copper homeostasis in Arabidopsis[J]. Plant Cell, 2009, 21(1): 347−361. doi: 10.1105/tpc.108.060137
[10] Stone J M, Liang X, Nekl E R, et al. Arabidopsis AtSPL14, a plant-specific SBP-domain transcription factor, participates in plant development and sensitivity to fumonisin B1[J]. Plant Journal, 2005, 41(5): 744−754. doi: 10.1111/tpj.2005.41.issue-5
[11] Chao L M, Liu Y Q, Chen D Y, et al. Arabidopsis transcription factors SPL1 and SPL12 confer plant thermotolerance at reproductive stage[J]. Molecular Plant, 2017, 10(5): 735−748. doi: 10.1016/j.molp.2017.03.010
[12] Unte U S, Sorensen A M, Pesaresi P, et al. SPL8, an SBP-box gene that affects pollen sac development in Arabidopsis[J]. Plant Cell, 2003, 15(4): 1009−1019. doi: 10.1105/tpc.010678
[13] Zhang Y, Schwarz S, Saedler H, et al. SPL8, a local regulator in a subset of gibberellin-mediated developmental processes in Arabidopsis[J]. Plant Molecular Biology, 2007, 63(3): 429−439. doi: 10.1007/s11103-006-9099-6
[14] Xing S, Salinas M, Hohmann S, et al. miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis[J]. Plant Cell, 2010, 22(12): 3935−3950. doi: 10.1105/tpc.110.079343
[15] Gou J, Debnath S, Sun L, et al. From model to crop: functional characterization of SPL8 in M. truncatula led to genetic improvement of biomass yield and abiotic stress tolerance in alfalfa[J]. Plant Biotechnology Journal, 2018, 16(4): 951−962. doi: 10.1111/pbi.2018.16.issue-4
[16] 李双, 苏艳艳, 王厚领, 等. 胡杨miR1444b在拟南芥中正调控植物抗旱性[J]. 北京林业大学学报, 2018, 40(4):1−9. Li S, Su Y Y, Wang H L, et al. Populus euphratica miR1444b positively regulates plants response to drought stress in Arabidopsis thaliana[J]. Journal of Beijing Forestry University, 2018, 40(4): 1−9.
[17] 姚琨, 练从龙, 王菁菁, 等. 胡杨PePEX11基因参与调节盐胁迫下拟南芥的抗氧化能力[J]. 北京林业大学学报, 2018, 40(5):19−28. Yao K, Lian C L, Wang J J, et al. PePEX11 functions in regulating antioxidant capacity of Arabidopsis thaliana under salt stress[J]. Journal of Beijing Forestry University, 2018, 40(5): 19−28.
[18] Sakuma Y, Maruyama K, Osakabe Y, et al. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression[J]. Plant Cell, 2006, 18(5): 1292−1309. doi: 10.1105/tpc.105.035881
[19] Strizhov N, Abraham E, Okresz L, et al. Differential expression of two P5CS genes controlling proline accumulation during salt-stress requires ABA and is regulated by ABA1, ABI1 and AXR2 in Arabidopsis[J]. Plant Journal, 1997, 12(3): 557−569.
[20] 李蕾蕾, 孙丰坤, 李天宇, 等. 白桦BpGT14基因启动子克隆及表达活性分析[J]. 北京林业大学学报, 2016, 38(7):16−24. Li L L, Sun F K, Li T Y, et al. Cloning and activity analysis of BpGT14 gene promoter in Betula platyphylla[J]. Journal of Beijing Forestry University, 2016, 38(7): 16−24.
[21] 张一南, 王洋, 张会龙, 等. 过表达胡杨PeRIN4基因拟南芥提高质膜H+-ATPase活性和耐盐性[J]. 北京林业大学学报, 2017, 39(11):1−8. Zhang Y N, Wang Y, Zhang H L, et al. Overexpression of PeRIN4 enhanced salinity tolerance through up regulation of PM H+-ATPase in Arabidopsis thaliana[J]. Journal of Beijing Forestry University, 2017, 39(11): 1−8.
[22] Jefferson R A, Kavanagh T A, Bevan M W. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants[J]. EMBO Journal, 1987, 6(13): 3901−3907. doi: 10.1002/embj.1987.6.issue-13
[23] Liu C, Guan M X, Hu X Q, et al. Complex regulatory network of Betula BplSPL8 in planta[J]. Journal of Forestry Research, 2017, 28(5): 881−889. doi: 10.1007/s11676-017-0372-0
[24] Yu X, Liu Y, Wang S, et al. CarNAC4, a NAC-type chickpea transcription factor conferring enhanced drought and salt stress tolerances in Arabidopsis[J]. Plant Cell Reports, 2016, 35(3): 613−627. doi: 10.1007/s00299-015-1907-5
[25] 牛素贞, 宋勤飞, 樊卫国, 等. 干旱胁迫对喀斯特地区野生茶树幼苗生理特性及根系生长的影响[J]. 生态学报, 2017, 37(21):7333−7341. Niu S Z, Song Q F, Fan W G, et al. Effects of drought stress on leaf physiological characteristics and root growth of the clone seedlings of wild tea plants[J]. Acta Ecologica Sinica, 2017, 37(21): 7333−7341.
[26] Yu N, Niu Q W, Ng K H, et al. The role of miR156/SPLs modules in Arabidopsis lateral root development[J]. Plant Journal, 2015, 83(4): 673−685. doi: 10.1111/tpj.12919
[27] Gao R, Wang Y, Gruber M Y, et al. miR156/SPL10 modulates lateral root development, branching and leaf morphology in Arabidopsis by silencing AGAMOUS-LIKE 79[J/OL]. Frontiers in Plant Science, 2017, 8 (2017−01−04) [2018−06−20]. https://doi.org/10.3389/fpls.2017.02226.
[28] 刘闯. 18个白桦SPLs基因的鉴定及BpSPL8基因的功能分析[D]. 哈尔滨: 东北林业大学, 2017. Liu C. Identification of 18 SPL gene family members and functional analysis of BpSPL8 in Betula platyphylla[D]. Harbin: Northeast Forestry University, 2017.
[29] Saini S, Sharma I, Kaur N, et al. Auxin: a master regulator in plant root development[J]. Plant Cell Reports, 2013, 32(6): 741−757. doi: 10.1007/s00299-013-1430-5
[30] Hao Y J, Wei W, Song Q X, et al. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants[J]. Plant Journal, 2011, 68(2): 302−313. doi: 10.1111/j.1365-313X.2011.04687.x
[31] Chen D, Richardson T, Chai S, et al. Drought-up-regulated TaNAC69-1 is a transcriptional repressor of TaSHY2 and TaIAA7, and enhances root length and biomass in wheat[J]. Plant and Cell Physiology, 2016, 57(10): 2076−2090. doi: 10.1093/pcp/pcw126
[32] Laplaze L, Benkova E, Casimiro I, et al. Cytokinins act directly on lateral root founder cells to inhibit root initiation[J]. Plant Cell, 2007, 19(12): 3889−3900. doi: 10.1105/tpc.107.055863
[33] Loutfy N, El-Tayeb M A, Hassanen A M, et al. Changes in the water status and osmotic solute contents in response to drought and salicylic acid treatments in four different cultivars of wheat (Triticum aestivum)[J]. Journal of Plant Research, 2012, 125(1): 173−184. doi: 10.1007/s10265-011-0419-9
[34] Ivanchenko M G, Muday G K, Dubrovsky J G. Ethylene-auxin interactions regulate lateral root initiation and emergence in Arabidopsis thaliana[J]. Plant Journal, 2008, 55(2): 335−347. doi: 10.1111/tpj.2008.55.issue-2
[35] Ruiz-Lozano J M, Aroca R, Zamarreno A M, et al. Arbuscular mycorrhizal symbiosis induces strigolactone biosynthesis under drought and improves drought tolerance in lettuce and tomato[J]. Plant Cell and Environment, 2016, 39(2): 441−452. doi: 10.1111/pce.v39.2
[36] Sanchez-Romera B, Ruiz-Lozano J M, Zamarreno A M, et al. Arbuscular mycorrhizal symbiosis and methyl jasmonate avoid the inhibition of root hydraulic conductivity caused by drought[J]. Mycorrhiza, 2016, 26(2): 111−122. doi: 10.1007/s00572-015-0650-7
[37] Rowe J H, Topping J F, Liu J, et al. Abscisic acid regulates root growth under osmotic stress conditions via an interacting hormonal network with cytokinin, ethylene and auxin[J]. New Phytologist, 2016, 211(1): 225−239. doi: 10.1111/nph.13882
[38] 赵婉莹, 于太飞, 杨军峰, 等. 大豆GmbZIP16的抗旱功能验证及分析[J]. 中国农业科学, 2018, 51(15):6−18. Zhao W Y, Yu T F, Yang J F, et al. Verification and analyses of soybean GmbZIP16 gene resistance to drought[J]. Scientia Agricultura Sinica, 2018, 51(15): 6−18.
-
期刊类型引用(24)
1. 张秀芸,伍文慧,梁英梅. 落叶松枯梢病在中国的适生性. 生态学报. 2024(07): 3027-3037 .  百度学术
百度学术
2. 葛婉婷,刘莹,赵智佳,张珅,李洁,杨桂娟,曲冠证,王军辉,麻文俊. 不同气候情景下黄心梓木在我国的潜在适生区预测. 林业科学. 2024(11): 63-74 .  百度学术
百度学术
3. 汤思琦,武扬,梁定东,郭恺. 未来气候变化下栎树猝死病菌在中国的适生性分析. 生态学报. 2023(01): 388-397 .  百度学术
百度学术
4. 刘璐璐,赵亮,蔺诗颖,冯建龙. 基于MaxEnt和GARP的阿蒙森海域南极磷虾(EUPHAUSIA SUPERBA)的分布区预测. 海洋与湖沼. 2023(02): 399-411 .  百度学术
百度学术
5. 唐雨薇,张晓龙,张雪云,吕佩锋,罗乐. 基于GIS与AHP分析法的单叶蔷薇生态适宜性评价. 绿色科技. 2023(13): 205-208+213 .  百度学术
百度学术
6. 王广平,李成,王书砚,刘超,杨君珑. 宁夏罗山青海云杉林空间分布特征研究. 农业科学研究. 2023(03): 10-15+23 .  百度学术
百度学术
7. 张惠惠,孟祥霄,林余霖,陈士林,黄林芳. 基于GMPGIS系统和MaxEnt模型预测人参全球潜在生长区域. 中国中药杂志. 2023(18): 4959-4966 .  百度学术
百度学术
8. 李盼畔,何旭诺,吴海荣,陈萍,刘明航,武目涛,王亚锋. 多年生豚草在中国的潜在分布预测. 植物检疫. 2022(04): 57-62 .  百度学术
百度学术
9. 李盼畔,何旭诺,左然玲,吕文刚,吴海荣. 4种蒺藜草属杂草在中国的潜在适生性预测. 杂草学报. 2022(02): 15-23 .  百度学术
百度学术
10. 林姗,陆兴利,王茹琳,李庆,王明田,郭翔,文刚. RCP8.5情景下气候变化对四川省猕猴桃溃疡病病菌地理分布的影响. 江苏农业科学. 2020(03): 124-129 .  百度学术
百度学术
11. 王华辰,朱弘,李涌福,伊贤贵,李蒙,南程慧,王贤荣. 中国特有植物雪落樱桃潜在分布及其生态特征. 热带亚热带植物学报. 2020(02): 136-144 .  百度学术
百度学术
12. 赵金鹏,王茹琳,刘原,陆兴利,王庆,郭翔,文刚,李庆. RCP4.5情景下四川省猕猴桃溃疡病菌适生性分析. 沙漠与绿洲气象. 2020(02): 137-143 .  百度学术
百度学术
13. 段义忠,王佳豪,王驰,王海涛,杜忠毓. 未来气候变化下西北干旱区4种扁桃亚属植物潜在适生区分析. 生态学杂志. 2020(07): 2193-2204 .  百度学术
百度学术
14. 陈爱莉,赵志华,龚伟,孔芬,张克亮. 气候变化背景下紫楠在中国的适宜分布区模拟. 热带亚热带植物学报. 2020(05): 435-444 .  百度学术
百度学术
15. 文雪梅,艾科拜尔·木哈塔尔,木巴来克·阿布都许科尔,阿不都拉·阿巴斯. 基于MaxEnt模型的新疆微孢衣属地衣生境适宜性评价. 武汉大学学报(理学版). 2019(01): 77-84 .  百度学术
百度学术
16. 常红,刘彤,王大伟,纪孝儒. 气候变化下中国西北干旱区梭梭(Haloxylon ammodendron)潜在分布. 中国沙漠. 2019(01): 110-118 .  百度学术
百度学术
17. 吕汝丹,何健,刘慧杰,姚敏,程瑾,谢磊. 羽叶铁线莲的分布区与生态位模型分析. 北京林业大学学报. 2019(02): 70-79 .  本站查看
本站查看
18. 陆兴利,罗伟,李庆,林姗,王茹琳,游超,郭翔,王明田. RCP2.6情景下四川省猕猴桃溃疡病菌潜在分布预测. 湖北农业科学. 2019(18): 49-54 .  百度学术
百度学术
19. 王蕾,罗磊,刘平,侯晓臣,邱琴,高亚琪,李曦光. 基于MaxEnt模型分析新疆特色林果区春尺蠖发生风险. 新疆农业科学. 2019(09): 1691-1700 .  百度学术
百度学术
20. 王茹琳,郭翔,李庆,王明田,游超. 四川省猕猴桃溃疡病潜在分布预测及适生区域划分. 应用生态学报. 2019(12): 4222-4230 .  百度学术
百度学术
21. 赵健,李志鹏,张华纬,陈宏,翁启勇. 基于MaxEnt模型和GIS技术的烟粉虱适生区预测. 植物保护学报. 2019(06): 1292-1300 .  百度学术
百度学术
22. 王奕晨,郑鹏,潘文斌. 运用GARP生态位模型预测福寿螺在中国的潜在适生区. 福建农林大学学报(自然科学版). 2018(01): 21-25 .  百度学术
百度学术
23. 邱靖,朱弘,陈昕,汤庚国. 基于DIVA-GIS的水榆花楸适生区模拟及生态特征. 北京林业大学学报. 2018(09): 25-32 .  本站查看
本站查看
24. 王野,陈磊,白云,张俊娥,刘红霞,田呈明. 云杉矮槲寄生遗传多样性的ISSR分析. 西北植物学报. 2017(11): 2153-2162 .  百度学术
百度学术
其他类型引用(20)



 下载:
下载: