BWB-assisted plus tree selection and deployment design for bulked progenies of the first-cycle Chinese pine seed orchard
-
摘要: 当前甘肃中湾油松良种基地正处于由第一育种周期向第二育种周期发展的关键转折时期,如何以前期改良成果为基础,结合现代林木育种的先进理论和技术,更高效地筛选和利用第二育种周期种质资源已经成为亟待解决的重要问题。本研究以甘肃中湾油松良种基地大规模初级种子园无谱系自由授粉混合子代群体为研究对象,详细阐述了基于种子园自由授粉混合子代的油松高育种周期优良种质资源选择、评价及配置利用策略等问题。结果表明,按照单株材积优势比的相对大小从中湾自由授粉混合子代中选出40个优良单株,其平均材积优势比为0.83;优良单株群体具有较高的遗传多样性,其平均多态性信息含量为0.620 2。基于SSR分子标记的亲缘分析结果表明,40个单株间亲缘关系不同,分属于23个同源组;为有效控制近交,进行了同源组内优良单株的二次选择,最终入选23个优良单株,并将之用作第二育种周期种子园的建园亲本。结合建园亲本材积优势比和亲缘分析的结果,制定了中湾第二育种周期种子园的分组、不平衡、完全、固定区组配置设计策略,兼顾了种子园子代材积遗传增益的提高和遗传多样性的维持。研究结果为油松高育种周期优良种质资源的高效选择及合理利用提供了新的思路,为缩短油松育种周期、提高油松育种效率奠定了基础。Abstract: Genetic improvement of Chinese pine in Zhongwan Improved Variety Base, Gansu Province of northwestern China, is in the key transition period from the first breeding cycle to the second breeding cycle. How to effectively select and utilize germplasm resources of the second breeding cycle, based on the previous improvement results and combined modern theories and technologies in tree breeding, has become important issues to be addressed immediately. Using large-scale no-peditree open-pollinated bulked progeny population from the first-generation seed orchard of Chinese pine in Zhongwan Improved Variety Base, Gansu Province, as study materials, this study elaborates issues about open-pollinated bulked progenies based elite germplasm resource selection, evaluation and utilization strategies for advanced cycle improvement of Chinese pine. Results showed that 40 superior individuals were selected based on the relative growth of stem volume, with the mean volume dominance ratio of 0.83. Population consisting 40 individuals had high levels of genetic diversity, with the mean polymorphism information content of 0.620 2. Results from SSR molecular markers based phylogenetic analysis showed that the 40 individuals had varying phylogenetic relationships, belonged to 23 homogenous groups. To effectively control inbreeding, the 40 individuals were conducted within-homogenous group selection and finally only 23 superior individuals were remained and used for parents of seed orchard in the second breeding cycle. Combined with the results of volume dominance ratios and phylogenetic analysis of these parents, seed orchard in the second breeding cycle of Zhongwan Improved Variety Base was suggested to adopt the grouped, unbalanced, complete, fixed block design, which had taken the increase of volume genetic gain and the maintenance of genetic diversity of seed orchrd progenies into consideration simultaneously. Results from this study will provide new insights into effective selection and rational utilization of elite germplasm resources in advanced breeding cycles of Chinese pine, and will lay the foundation for shortening breeding cycle and increasing breeding efficiency in genetic improvement of Chinese pine.
-
Keywords:
- Chinese pine /
- BWB /
- germplasm selection /
- phylogenetic analysis /
- deployment design
-
近交是树种遗传改良过程中需考虑的重要问题之一[1-2],为有效控制近交,树种高轮次遗传改良过程中多通过控制授粉获得亲缘关系清楚的全同胞家系,并从中选择少数亲缘关系较远的优良子代个体,构建高轮次改良的繁育群体。然而,从控制授粉到获得子代测定结果需要较长的时间,且可供选择的组合数和全同胞家系规模有限,限制了发现遗传品质更优子代个体的可能性。从种子园自由授粉子代中选择优良个体,减少了繁琐的控制授粉环节、增加了可供选择的组合数和家系规模,但由于个体间亲缘关系不清楚,存在近交的可能,难以直接应用于高轮次繁育群体选择过程中。
分子标记技术的发展使得在DNA水平上直接鉴别个体间的亲缘关系成为可能[3-5]。BWB(Breeding Without Breeding)是近几年发展起来的育种策略,2009年El-Kassaby等[6-7]将该技术应用到树木育种领域,指出可以直接利用分子标记技术鉴别自由授粉子代个体间的亲缘关系。当前,BWB策略已在多个树种的遗传改良过程中得到了验证和应用[8-9]。随着分子标记技术在鉴别子代亲缘关系中的有效利用,直接从种子园自由授粉子代中选择优良单株,辅以基于分子标记亲缘鉴别的BWB育种策略,可显著缩短育种周期;并可从包括所有可能亲本组合且规模庞大的自由授粉子代群体中获得最优组合及个体,提高下一育种周期无性系群体的育种值。此外,分子标记可以检测高改良周期种子园无性系间的相对亲缘关系[5],在无性系配置设计中可以根据亲缘关系远近,合理配置无性系,以更有效控制近交率,提高远缘配置几率,拓展子代遗传多样性基础。
油松(Pinus tabuliformis)作为中国最重要的针叶树种之一,在北方山区的经济发展和生态建设中具有重要地位[10-11]。甘肃庆阳的中湾油松良种基地油松种子园正处于由初级向高轮次发展的关键转折期。本文将以正宁油松初级种子园大规模自由授粉混合子代群体为研究对象,通过评价种子园自由授粉混合子代家系优良单株的材积生长优良度;分析SSR分子标记对优良单株亲缘关系分析的有效性;制定基于亲缘关系分析的子代无性系配置设计策略等过程选择表型最优个体,鉴别其相对亲缘关系,并根据亲缘关系制定无性系配置设计。以期为油松高育种周期优良种质资源的高效选择提供新的思路,为促进分子标记在油松高育种周期种子园建设中的应用提供参考。
1. 材料与方法
1.1 油松第二育种周期基本群体
中湾油松良种基地位于甘肃省正宁县,地理位置为103°30′ E、35°30′ N,地处油松中心分布区,海拔1 600 m左右,坡度范围为5°~25°。土壤肥力中等,pH值变动范围为7.5~8.1。研究地位于北温带大陆性季风气候区,平均年降水量在600 mm左右,年均温8.3 ℃,平均无霜期163 d,年均日照2 447 h。该良种基地已用油松初级种子园无谱系自由授粉混合种子在环境条件相对一致的林区于1991年营造了24.53 hm2人工林,林内株行距2 m×2 m,管理条件基本一致,总体生长良好,现保存58 033个油松单株,本研究将以人工林为油松第二育种周期的基本群体。
1.2 油松第二育种周期优良单株选择
2012年测定了基本群体内油松单株的树高(m)和胸径(cm),按照实验形数法[12]计算单株材积(m3)。根据单株材积优势比的相对大小(Rd)进行第二育种周期优良单株选择,单株材积优势比的计算公式为:
Rd=(VPT−¯V)/¯V 式中:VPT代表优良单株材积,V代表所有子代单株材积均值。
1.3 优良单株间的相对亲缘关系分析
以筛选出的优良单株群体为实验样本,用CTAB法[13]提取油松基因组DNA。以油松基因组DNA为模板,用前期筛选的11对油松多态性SSR引物[14-15]进行PCR扩增。所用引物由生工生物工程(上海)有限公司合成。PCR扩增体系为20 μL, 其中包括基因组DNA 1 μL、正反向引物各1 μL、2×PCR Master Mix(北京艾德莱生物科技有限公司)10 μL和ddH2O 7 μL。PCR扩增程序首先为95 ℃预变性5 min,然后是35个循环的95 ℃变性30 s、51~58 ℃退火45 s、72 ℃延伸30 s,最后是72 ℃继续延伸10 min,4 ℃保存。PCR扩增得到的产物在聚丙烯酰胺凝胶(浓度为8%)上进行基因型分型,实验操作方法按说明书上的步骤进行。基因型分型结果用PowerMarker软件(v 3.25)计算等位基因频率和基于等位基因频率的遗传距离,并依据遗传距离进行聚类分析(UPGMA法),用FigTree软件(v 1.4.2)查看聚类结果。根据遗传距离的聚类结果分析群体内单株间的相对亲缘关系。
1.4 优良单株群体的遗传多样性分析
用PowerMarker软件分析第二育种周期优良单株群体的遗传多样性。遗传多样性分析参数包括主要观测杂合度(observed heterozygosity, Ho)、期望杂合度(expected heterozygosity, He)、固定指数(fixation index, F)、多态信息含量(polymorphism information content, PIC)等。
2. 结果与分析
2.1 油松第二育种周期优良单株选择
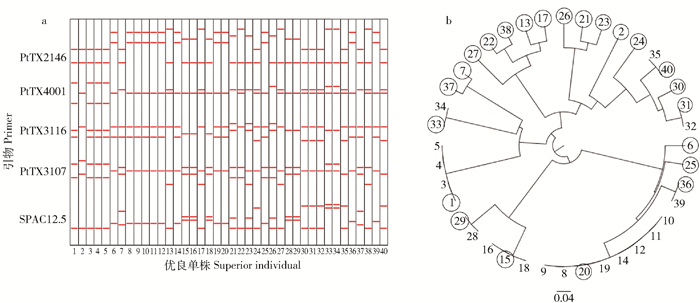
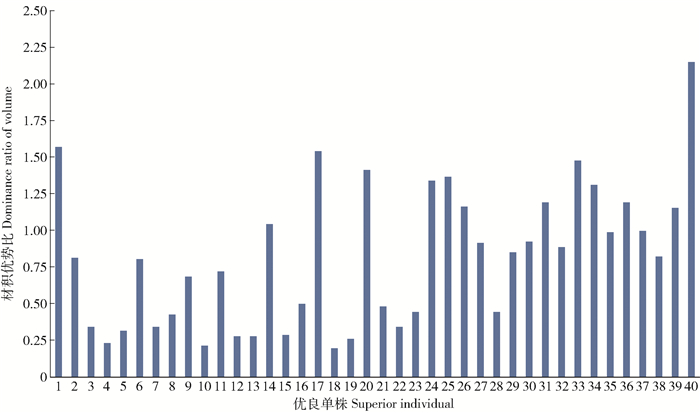
在中湾油松良种基地初级种子园无谱系自由授粉混合子代人工林内,以单株材积优势比为参考指标进行油松第二育种周期优良单株选择。从选择结果可以看出,以单株材积优势比≥0.2作为选择标准,在中湾油松良种基地初级种子园自由授粉混合子代中共选出40个优良单株,入选率为0.069%,其中40号和18号单株的材积优势比处于入选单株的最高和最低水平,其材积优势比分别为2.15和0.20,优良单株群体的平均材积优势比为0.83(图 1)。
2.2 油松第二育种周期优良单株的亲缘分析
在11个SSR位点上分析中湾油松良种基地初级种子园无谱系自由授粉子代人工林内优良单株间的亲缘关系。指纹图谱检测结果显示,筛选出的40个优良单株间在引物扩增位点上的多态性程度不同,部分单株间在引物扩增位点上存在多态性,如2、7、10、17、28、32号单株间;部分单株间在引物扩增位点上不存在多态性,如8、9、10、11、12号单株间(图 2a)。亲缘分析结果进一步表明,40个单株共来自23个同源组。各同源组内单株数目不同,其中数目最多的同源组包含8个单株,分别为8、9、10、11、12、14、19和20号;仅包含1个单株的同源组共15个;包含2个单株的同源组5个;包含3和4个单株的同源组各1个(图 2b)。同源组间相对亲缘关系不同,例如由28、29号单株构成的同源组与由15、16、18号单株构成的同源组间的亲缘关系较近,而这两个同源组与其余同源组间具有相对较远的亲缘关系;同理,由13、17、22、27、38号单株构成的同源组具有较近的亲缘关系,但这些亲缘组与其他单株构成同源组之间具有相对较远的亲缘关系;各同源组间的相对亲缘关系分析结果详见图 2b。
2.3 甘肃正宁第二育种周期油松优良单株构成群体的遗传多样性测定
如图 3所示,由40个单株、23个同源组构成的优良单株群体,其在11个SSR位点上的基因型数变化在4.000 0和10.000 0之间,平均值为6.090 9(图 3a);等位基因数变化在3.000 0和11.000 0之间,平均值为5.545 5(图 3a),基因型数和等位基因数均较丰富。期望杂合度的平均值为0.669 0、观测杂合度的平均值为0.561 4(图 3b),观测杂合度和期望杂合度间有一定的差距。固定指数变化在-0.267 6和0.853 2之间,平均值为0.173 2(图 3c),说明该群体内存在一定程度的近交几率。多态信息含量变化在0.397 7和0.767 1之间,平均值为0.620 2(图 3d),说明群体的遗传多样性水平较高。
2.4 油松第二育种周期种子园无性系配置设计
当前,油松的育种目标以用材为主,材积生长无疑是育种工作者更关心的性状。本研究中筛选出的40个优良单株由于具有材积生长优势,且遗传多样性水平较高,是新一周期种子园建园亲本的重要来源。但从亲缘分析和遗传多样性分析结果中可以看出,40个单株共来自23个同源组,40个单株构成的群体存在一定程度的近交几率。为维持群体遗传多样性并有效控制近交,进行了优良单株的二次选择,即根据单株材积优势比的相对大小,对单株数≥2的同源组进行了组内选择,同一个同源组内仅保留材积优势比最大的1株,最终除去17个单株,保留23个优良单株(图 2b中用圆圈标出)。
将最终保留的23个油松优良单株用作建园亲本,通过无性嫁接的方式建成23个无性系,无性系的编号与其来源单株的编号相同。配置设计前首先根据无性系间的亲缘关系将23个无性系分为6亲缘组,同组内无性系间亲缘关系近,组间无性系亲缘关系远,如6、25、36、20号无性系分到同一亲缘组,15、29号无性系分到同一亲缘组,其余分组情况详见表 1,亲缘组内无性系数目的变动范围为2~5个。
表 1 油松第二育种周期种子园亲缘组和配置组分组情况Table 1. Phylogenetic group and deployment group in the second breeding cycle seed orchard of Chinese pine组别
Group无性系
编号
Clone No.无性系
编号
Clone No.无性系
编号
Clone No.无性系
编号
Clone No.无性系
编号
Clone No.1 6 25 36 20 2 15 29 3 1 33 37 7 4 27 22 38 13 17 5 26 21 23 6 2 24 40 30 31 配置设计时采用分组、不平衡、完全、固定区组设计(图 4)。在每个亲缘组内选择1个无性系构成1个配置组,1个配置组由亲缘关系较远的6个无性系构成(图 4中一个方框内的无性系构成一个配置组)。所有无性系全部配置完全需5个配置组,亲缘组内无性系数目不足5的用组内材积优势比最大的无性系补充;最后增加1个配置组,由各亲缘组内材积优势比最大的无性系构成(图 4中右下角方框内无性系构成的配置组)。6个配置组构成1个完全区组(图 4中由灰色底纹标示),同一区组内最优无性系(各亲缘组内材积优势比最大的无性系)、较优无性系(除最优无性系和较差无性系以外的无性系)和较差无性系(各亲缘组内材积优势比最小的无性系)数目分别为19(52.78%,图 4中加下划线的无性系)、11(30.56%)和6(16.67%,图 4中用斜体标明的无性系),实现了无性系间的不平衡配置。各区组内无性系的相对位置保持固定不变。图 4显示的是4个区组的配置设计图。
3. 讨论
3.1 油松第二育种周期种子园建园亲本选择策略
本研究中的基本群体为中湾油松初级种子园自由授粉混合子代,群体种植规模大,其父母本均未知。以群体内单株材积的相对优势比为参考指标进行油松第二育种周期种子园建园亲本选择,从选择结果可以看出,入选建园亲本的平均单株材积优势比为0.83(图 1),显著高于在山西隰县油松初级种子园自由授粉子代测定林中筛选出的建园亲本的平均单株材积优势比0.25[14]。这可能是因为与自由授粉子代测定林相比,本研究中的自由授粉子代林种植规模大,包含了更多测定家系,更接近油松初级种子园所有无性系随机交配子代,群体遗传多样性水平较高,选择潜力更大。遗传多样性分析结果表明,来自种子园大规模自由授粉子代人工林的优良单株群体内的基因型数和等位基因数位点较丰富,平均值分别为6.090 9和5.545 5(图 3a、3b);群体的遗传多样性水平较高,平均多态信息含量为0.620 2(图 3d),要高于从山西隰县种子园自由授粉子代测定林选出的优良单株群体[14]。因此,本研究中筛选出的优良单株在具有较高材积遗传增益的同时,还具有较高的遗传多样性,在今后油松遗传改良过程中,可以将高育种周期种子园繁育群体的选择范围扩展到大规模种植的油松种子园自由授粉子代人工林内,从而显著提高下一育种周期无性系群体的育种值,将目标性状的遗传增益更大化,提高改良的效果。
3.2 基于SSR分子标记的子代亲缘分析的有效性
20世纪80年代发展起来的SSR分子标记技术以其中性进化、位点丰富、在基因组中均匀分布、共显性、稳定、可重复等优点,广泛应用于品种鉴定、亲缘分析和标记辅助选择等研究领域。本试验利用筛选出的11对扩增条带稳定、清晰、可重复的油松多态性SSR引物[14],对入选优良单株进行了标记位点的扩增,结果显示,这些多态性引物可用于区分来自油松种子园自由授粉子代人工林中的不同油松单株。
近交会使空籽率增加,种子产量下降;即使得到饱满种子,其活力也较低[1-2]。为有效控制近交,高育种周期种子园往往将谱系清楚的优良全同胞家系作为建园材料,但控制授粉过程费时费力,明显降低了改良的效率。本试验用11对多态性SSR引物鉴别正宁油松初级种子园自由授粉混合子代群体内入选优良单株之间的相对亲缘关系。鉴定结果表明,这些单株间在所选引物的扩增位点上存在不同程度的多态性(图 2a)。单株间具有不同程度的亲缘关系,发现多个单株同属于一个同源组的情形,40个优良单株分属于23个同源组,如8、9、10、11、12、14、19和20号单株属于同一同源组(图 2a、2b)。这一方面说明该同源组确实具有显著的材积生长优势,另一方面说明在种子园自由授粉混合子代中进行优良基因型选择时存在选到相同同源组的几率,因此选择时入选单株数目应大于实际需要单株数目,然后根据亲缘分析结果进行同源组内单株二次选择。以入选的23个单株为亲本,形成23个无性系,根据无性系间的相对亲缘关系,可将所有测定无性系分为5个亲缘组,组内无性系间亲缘关系近,组间亲缘关系远。例如,由28、29号单株构成的无性系与由15、16、18号单株构成的无性系其亲缘关系较近,分到同一亲缘组;同理,13、17、22、27、38号单株构成的无性系分到同一亲缘组(表 1)。将SSR分子标记技术应用于亲缘关系完全未知的优良单株的亲缘分析过程中,虽不能鉴定各单株的详细亲本来源,却能在分子水平上明确不同单株之间亲缘关系的相对远近,在高育种周期改良中,通过把亲缘关系远近不同的单株配置在合理的位置,可以避免近交。因此,在种子园自由授粉子代人工林中选择高育种周期种子园的繁殖群体,辅以基于分子标记的相对亲缘关系分析策略,可省去繁琐的控制授粉环节,显著缩短育种周期,提高改良的效率、效益和效果。
3.3 基于亲缘分析的无性系配置设计
种子园是为生产提供优良种子或繁殖材料的繁殖群体[16-17]。营建种子园的最重要目的就是在维持一定遗传多样性的情况下最大限度地提高目标性状的遗传增益。有效群体大小、交配系统、开花物候等因素都直接或间接影响遗传多样性或遗传增益,是种子园配置设计时需要考虑的问题[18]。正宁油松初级种子园采取的是顺序错位配置设计,由于初级种子园建园无性系数目较多、亲缘关系较远,采用这种配置设计是可行的。高育种周期种子园中包含的无性系数量较少,且无性系来源多样化,近交几率增大,因此配置设计也更加复杂[18]。
高育种周期种子园无性系数量少,且优良,为减少自交和管理方便,通常采用固定配置[19]。同时,为了将遗传距离适中的无性系配合一起,增加子代的遗传基础,设计中采用固定配置更能实现该目的。基于材积相对优势比的大小和亲缘关系分析的结果,正宁第二代种子园采用分组、不平衡、完全固定区组设计,按照无性系间亲缘关系,将23个无性系分为6个配置组,每组包含6个来自不同亲缘组的无性系,每个无性系1个单株,组内无性系间亲缘关系较远;每个区组内来自最优无性系、较优无性系和较差无性系的数目分别占区组内无性系总数的52.78%、30.56%和16.67%;1个区组包含所有测定无性系;所有区组无性系的排列完全相同(图 4)。采用分组固定配置,有效促进了远缘交配,使有近缘关系的优良子代无性系在改良代种子园中得以有效控制利用。同时,与前人研究方法相同[20-21],本研究采用不平衡区组配置,使区组平均材积优势比由平衡设计的1.00提高到1.76,在遗传多样性不变的前提下将材积遗传增益提高到了一个更高层次。
针对油松高育种周期遗传改良中的种质选择和利用问题,本研究冲破了高育种周期种质应来源于种子园无性系或种子园自由授粉、控制授粉子代测定林内优良家系的束缚,首次提出油松高育种周期的种质可来源于大规模种子园自由授粉子代人工林中的优良单株,并从材积生长、遗传多样性水平等方面分析了将这些单株用作高育种周期优良种质资源的优势,为油松遗传改良的种质来源提供了新的思路。同时,本研究利用SSR分子标记对来自种子园子代人工林中的优良单株进行了基于遗传距离的相对亲缘关系分析,解决了单株之间亲缘关系不清楚,无法直接应用于高育种周期种子园的问题。最后,基于优良单株间材积生长优势比的相对大小及亲缘关系远近,本研究提出了油松高育种周期种子园的分组、不平衡、固定配置设计策略,为油松高育种周期种子园的配置设计提供了新的思路。总之,本研究的结果为油松的高轮次遗传改良提供了重要借鉴,将有效促进油松遗传改良向着效率更高、效益更大、效果更好的方向持续发展。
-
表 1 油松第二育种周期种子园亲缘组和配置组分组情况
Table 1 Phylogenetic group and deployment group in the second breeding cycle seed orchard of Chinese pine
组别
Group无性系
编号
Clone No.无性系
编号
Clone No.无性系
编号
Clone No.无性系
编号
Clone No.无性系
编号
Clone No.1 6 25 36 20 2 15 29 3 1 33 37 7 4 27 22 38 13 17 5 26 21 23 6 2 24 40 30 31 -
[1] NAMKOONG G. Inbreeding effects on estimation of genetic additive variance[J]. Forest Science, 1966, 12(1): 8-13. https://www.ingentaconnect.com/content/saf/fs/1966/00000012/00000001/art00004
[2] GRIFFIN A R, LINDGREN D. Effect of inbreeding on production of filled seed in Pinus radiata-experimental results and a model of gene action[J]. Theoretical and Applied Genetics, 1985, 71(2): 334-343. doi: 10.1007/BF00252077
[3] NIELSEN R, MATTILA D K, CLAPHAM P J, et al. Statistical approaches to paternity analysis in natural populations and applications to the North Atlantic humpback whale[J]. Genetics, 2001, 157(4): 1673-1682. http://d.old.wanfangdata.com.cn/OAPaper/oai_pubmedcentral.nih.gov_1461600
[4] JONES A G, AREREN W R. Methods of parentage analysis in natural populations[J]. Molecular Ecology, 2003, 12(10): 2511-2523. doi: 10.1046/j.1365-294X.2003.01928.x
[5] MASSAH N, WANG J L, RUSSELL J H, et al. Genealogical relationship among members of selection and production populations of yellow cedar (Callitropsis nootkatensis [D. Don] Oerst.) in the absence of parental information[J]. Journal of Heredity, 2010, 101(2): 154-163. doi: 10.1093/jhered/esp102
[6] EL-KASSABY Y A, LSTIBǓ REK M. Breeding without breeding[J]. Genetics Research, 2009, 91(2): 111-120. doi: 10.1017/S001667230900007X
[7] EL-KASSABY Y A, CAPPA E P, LIEWLAKSANEEYANAWIN C, et al. Breeding without breeding: is a complete pedigree necessary for efficient breeding[J/OL]. PLoS ONE, 2011, 6(10): e25737.http://dx.doi.org/10.1371/journal.pone.0025737. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3185014/?report=abstract
[8] WANG X R, TORIMARU T, LINDGREN D, et al. Marker-based parentage analysis facilitates low input 'breeding without breeding' strategies for forest trees[J]. Tree Genetics & Genomes, 2010, 6(2): 227-235. http://www.wanfangdata.com.cn/details/detail.do?_type=perio&id=3934db0d04ab4bdf296afb8d3379b172
[9] ZHAO P, ZHANG S Z, WOESTE K. Genotypic data changes family rank for growth and quality traits in a black walnut (Juglans nigra L.) progeny test[J]. New Forests, 2013, 44(3): 357-368. doi: 10.1007/s11056-012-9343-7
[10] CHEN K M, ABBOTT R J, MILNE R I, et al. Phylogeography of Pinus tabulaeformis Carr. (Pinaceae), a dominant species of coniferous forest in northern China[J]. Molecular Ecology, 2008, 17(19): 4276-4288. doi: 10.1111/j.1365-294X.2008.03911.x
[11] YUAN H W, LI Z X, FANG P, et al. Variation and stability in female strobili production of a first-generation clonal seed orchard of Chinese pine (Pinus tabuliformis)[J]. Silvae Genetica, 2014, 63(1/2): 41-47. http://www.wanfangdata.com.cn/details/detail.do?_type=perio&id=c5846e1fc292d59f053a07b773de509f
[12] 张新波, 李悦, 袁虎威, 等.山西油松天然林分21年子代生长性状遗传变异研究[J].北京林业大学学报, 2014, 36(3): 104-109. doi: 10.13332/j.cnki.jbfu.2014.03.016 ZHANG X B, LI Y, YUAN H W, et al. Genetic variations of growth traits in a 21-year-old stand progeny of Shanxi natural Pinus tabuliformis forests[J]. Journal of Beijing Forestry University, 2014, 36(3): 104-109. doi: 10.13332/j.cnki.jbfu.2014.03.016
[13] POREBSKI S, BAILEY L G, BAUM B R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components[J]. Plant Molecular Biology Reporter, 1997, 15(1): 8-15. doi: 10.1007/BF02772108
[14] 袁虎威, 梁胜发, 符学军, 等.山西油松第二代种子园亲本选择与配置设计[J].北京林业大学学报, 2016, 38(3): 47-54. doi: 10.13332/j.1000-1522.20150370 YUAN H W, LIANG S F, FU X J, et al. Parental selection and deployment design in the second-generation seed orchard of Chinese pine in Shanxi Province[J]. Journal of Beijing Forestry University, 2016, 38(3): 47-54. doi: 10.13332/j.1000-1522.20150370
[15] 张冬梅, 杨娅, 沈熙环, 等.油松SSR-PCR引物筛选及反应体系的建立[J].北京林业大学学报, 2007, 29(2): 13-17. http://j.bjfu.edu.cn/article/id/9270 ZHANG D M, YANG Y, SHEN X H, et al. Selection of primers and establishment of SSR-PCR reaction system on Pinus tabuliformis Carr.[J]. Journal of Beijing Forestry University, 2007, 29(2): 13-17. http://j.bjfu.edu.cn/article/id/9270
[16] BURCZYK J, CHALUPKA W. Flowering and cone production variability and its effect on parental balance in a Scots pine clonal seed orchard[J]. Annals of Forest Science, 1997, 54(2): 129-144. doi: 10.1051/forest:19970201
[17] KANG K S, LINDGREN D. Fertility variation among clones of Korean pine (Pinus koraiensis S. et Z.) and its implications on seed orchard management[J]. Forest Genetics, 1999, 6(3): 191-200.
[18] HODGE G R, WHITE T L. Advanced-generation wind-pollinated seed orchard design[J]. New Forests, 1993, 7(3): 213-236. doi: 10.1007/BF00127387
[19] WHITE T L, ADAMS W T, NEALE D B. Forest genetics[M]. Cambridge: CABI Publishing, 2007.
[20] LINDGREN D, DANUSEVICIUS D, ROSVALL O. Unequal deployment of clones to seed orchards by considering genetic gain, relatedness and gene diversity[J]. Forestry, 2009, 82(1): 17-28. doi: 10.1093/forestry/cpn033
[21] WENG Y H, PARK Y S, LINDGREN D. Unequal clonal deployment improves genetic gains at constant diversity levels for clonal forestry[J]. Tree Genetics & Genomes, 2012, 8(1): 77-85. http://www.wanfangdata.com.cn/details/detail.do?_type=perio&id=9fe802aec60822f11577ec4f806db0fd
-
期刊类型引用(12)
1. 孙凡,马彦广,刘占民,杨博宁,王辉丽,李伟. 油松高世代种子园亲本选择策略研究. 北京林业大学学报. 2024(04): 28-39 .  本站查看
本站查看
2. 吕寻,李万峰,胡勐鸿,戴小芬,成红梅,委霞. 日本落叶松种子园和优树自由授粉家系选择与利用研究. 西南林业大学学报(自然科学). 2024(03): 1-9 .  百度学术
百度学术
3. 冯健,张金博,杨圆圆,杜超群,徐柏松,曹颖,姚飞. 基于生长性状和SSR遗传多样性分析的红松第二代优树选择研究. 西南林业大学学报(自然科学). 2024(04): 1-7 .  百度学术
百度学术
4. 胡勐鸿,吕寻,戴小芬,李宗德,李万峰. 日本落叶松无性系种子园和优树半同胞家系苗期比较. 东北林业大学学报. 2024(12): 10-17 .  百度学术
百度学术
5. 向华,向文明,向明. 我国用材林优树选择技术研究进展. 湖南林业科技. 2021(02): 89-96 .  百度学术
百度学术
6. 康向阳. 林木遗传育种研究进展. 南京林业大学学报(自然科学版). 2020(03): 1-10 .  百度学术
百度学术
7. 邓乐平,黄婷,王哲,吴惠姗,李晓华,廖仿炎,李义良,郭文冰,赵奋成. 湿地松改良种子园无性系的遗传评价及新一轮育种亲本选择. 林业与环境科学. 2020(04): 1-7 .  百度学术
百度学术
8. 杜超群,赵虎,袁慧,侯义梅,朱于勤,许业洲. 日本落叶松种子园母树生长及种实性状评价. 森林与环境学报. 2019(01): 32-36 .  百度学术
百度学术
9. 王芳,王元兴,王成录,张伟娜,刘卫胜,陆志民,杨雨春. 红松优树半同胞子代家系生长、结实及抗病虫能力的变异特征. 应用生态学报. 2019(05): 1679-1686 .  百度学术
百度学术
10. 金星,于忠峰,苗海伟,于国斌,朱瑞,张丽杰. 辽宁地区油松花粉形态及生活力的测定. 分子植物育种. 2019(15): 5115-5119 .  百度学术
百度学术
11. 康向阳. 关于林木育种策略的思考. 北京林业大学学报. 2019(12): 15-22 .  本站查看
本站查看
12. 苗禹博,朱晓梅,李志娟,贾凤岭,李伟. 不同世代樟子松育种资源遗传评价. 北京林业大学学报. 2017(12): 71-78 .  本站查看
本站查看
其他类型引用(4)



 下载:
下载: